Biological Evaluation and Molecular Docking of Protocatechuic Acid from Hibiscus sabdariffa L. as a Potent Urease Inhibitor by an ESI-MS Based Method
Abstract
:1. Introduction
2. Results
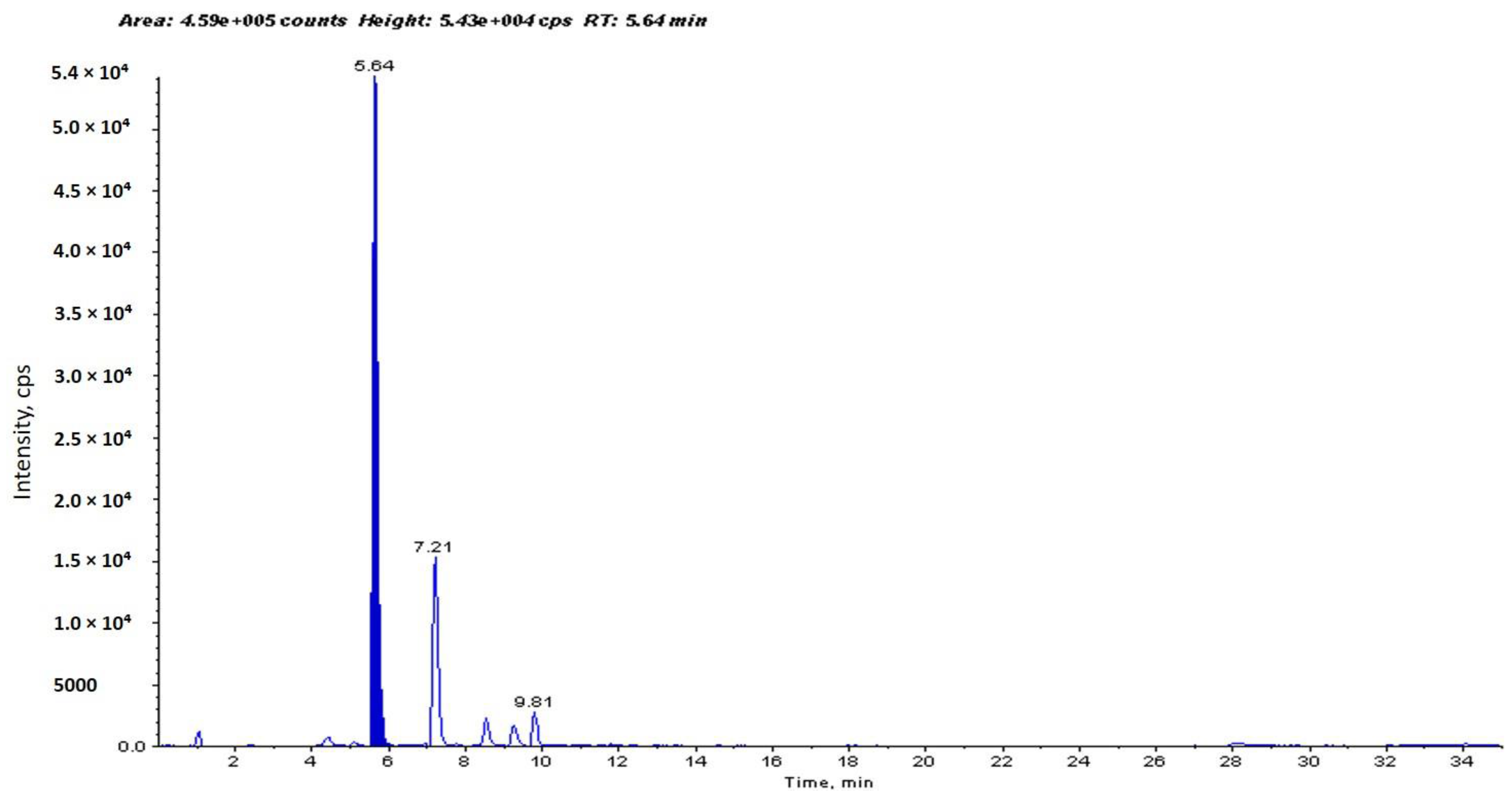
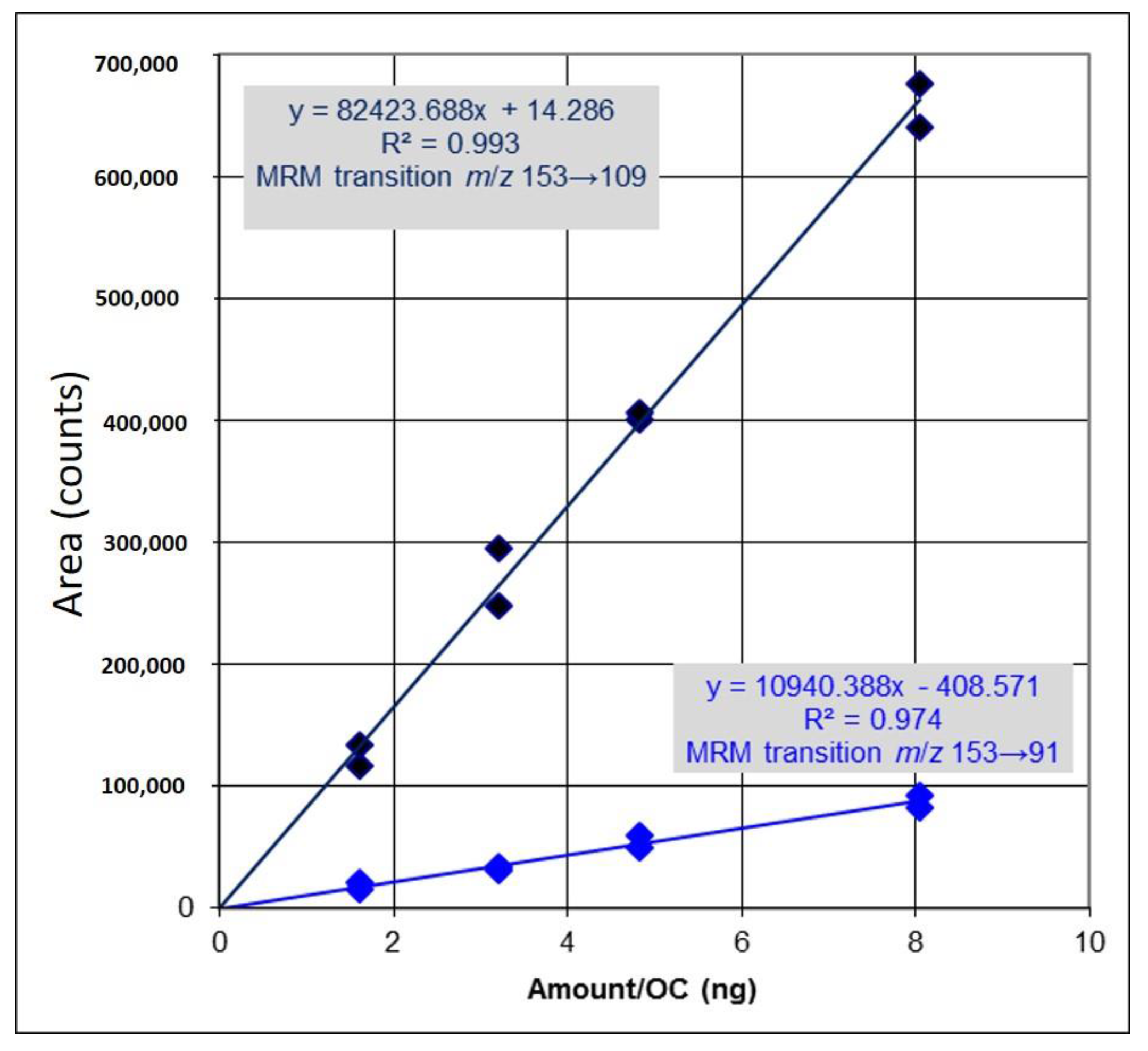
2.1. Identification and Determination of Concentration of PCA in H. sabdariffa
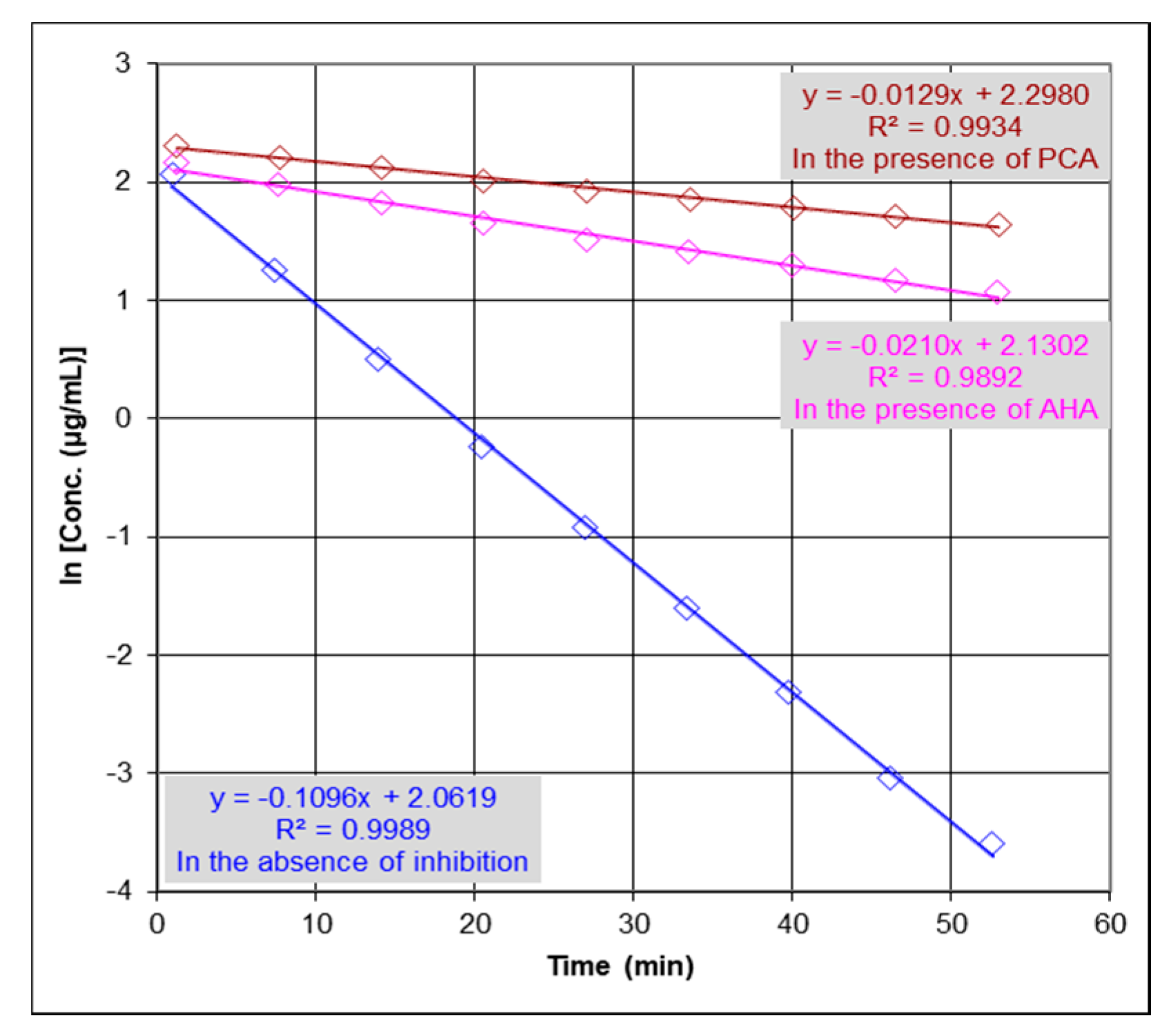
2.2. Urease Inhibition Studies and Determination of IC50
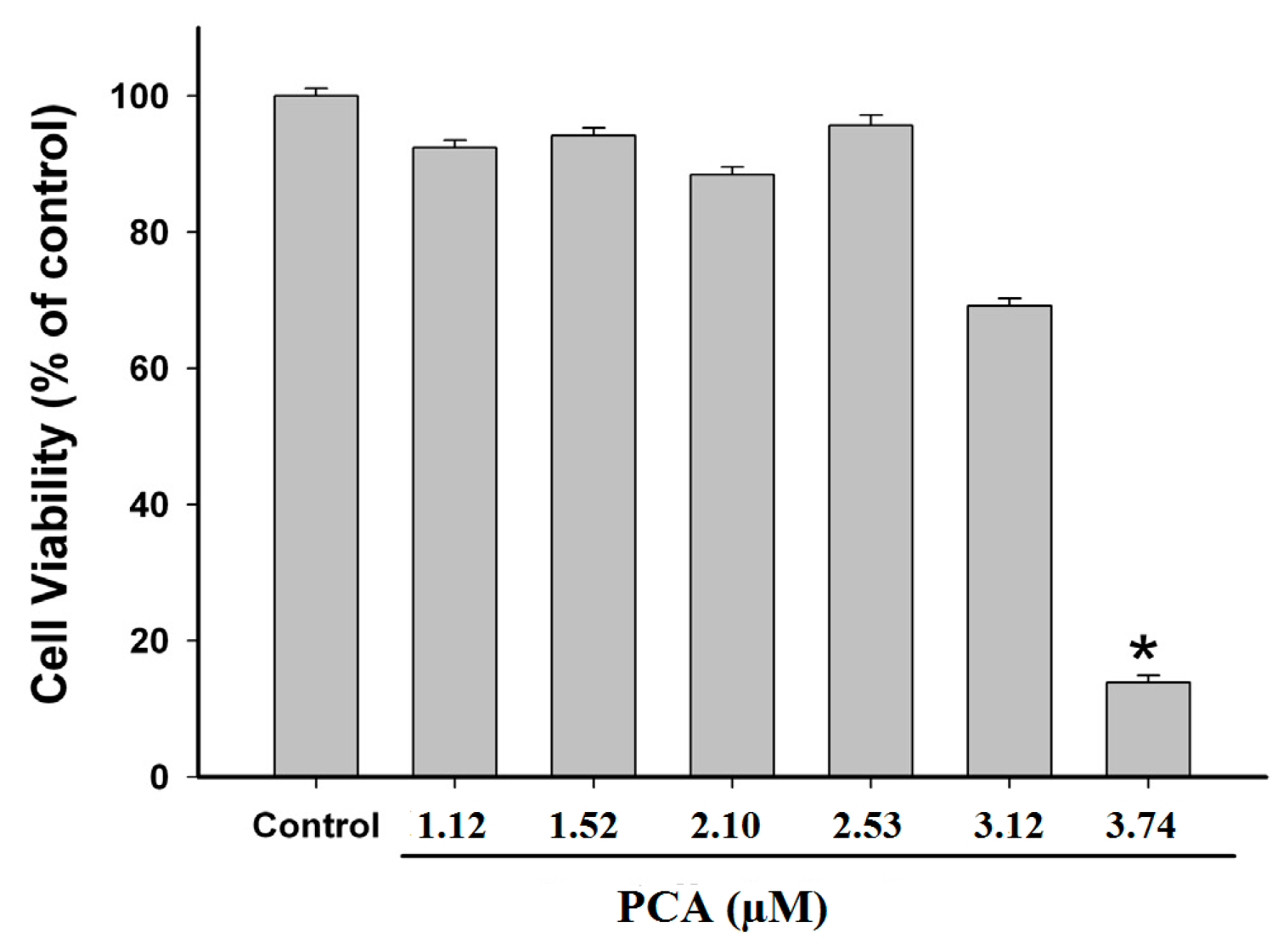
2.3. Effect of PCA on GES-1 Viability
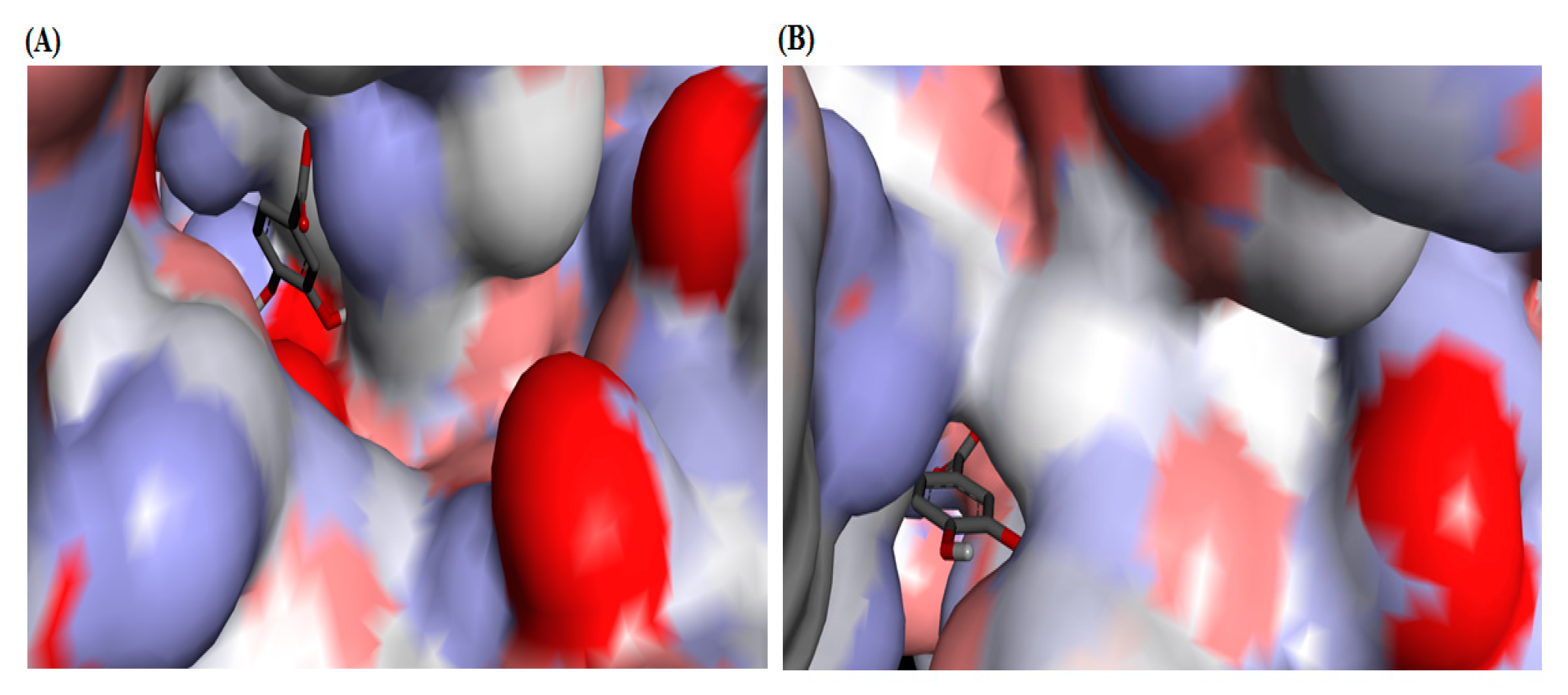
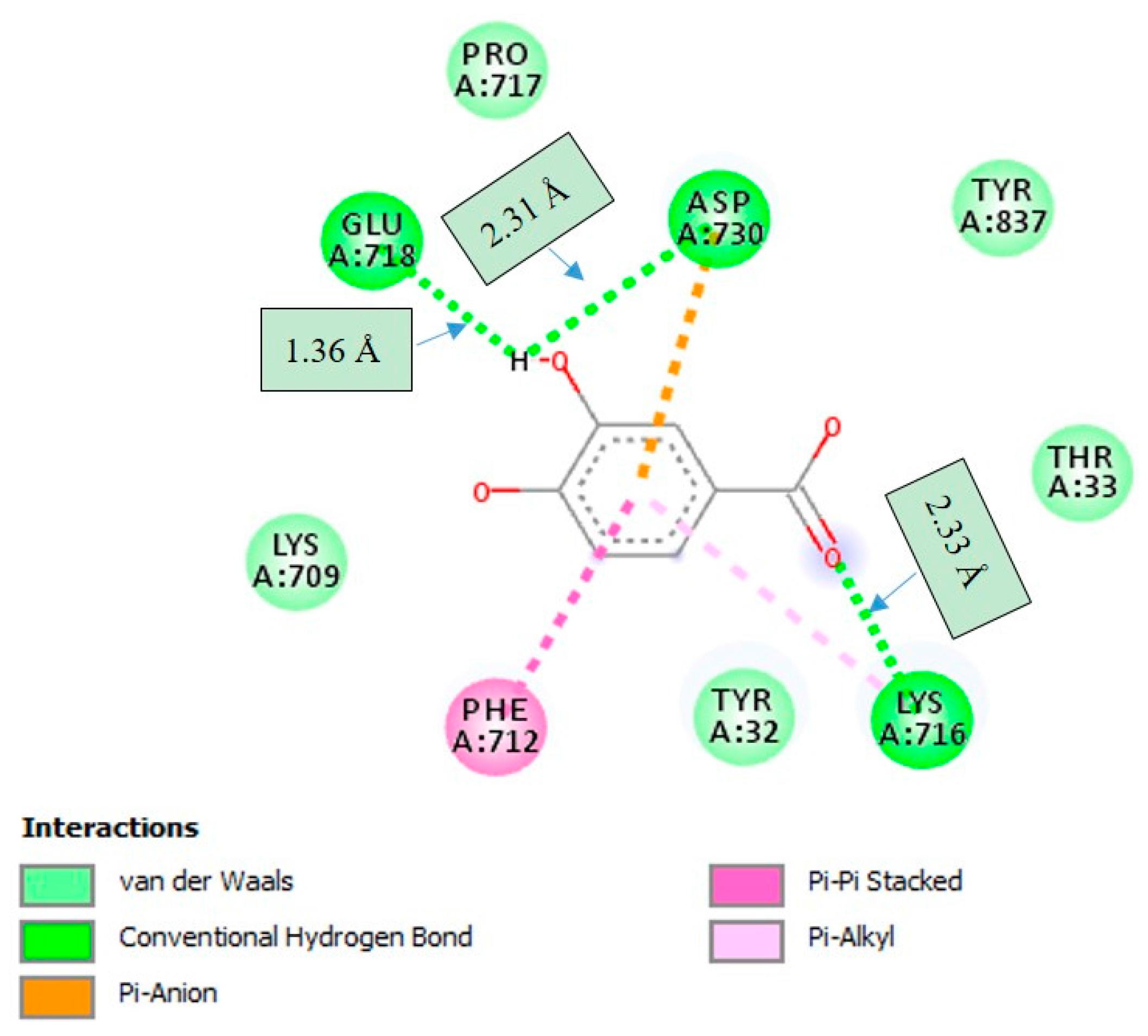
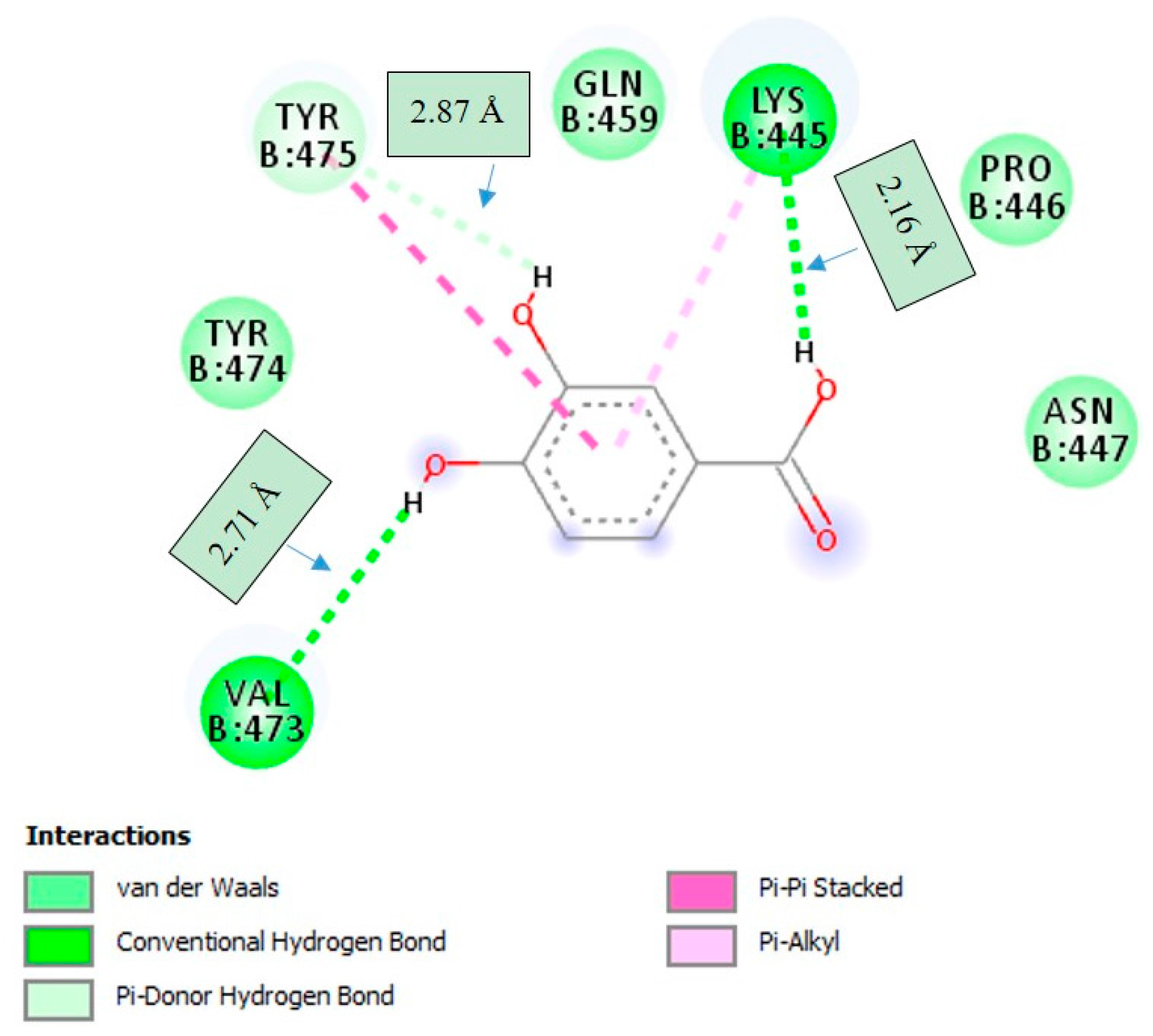
2.4. Molecular Docking Studies
3. Discussion
4. Materials and Methods
4.1. Identification and Determination of Concentration of PCA in H. sabdariffa
4.2. Enzyme, Substrate, Inhibitors, and Reagents
4.3. ESI-MS
4.4. Anti-Urease Activity by ESI-MS-Based Assay
4.5. Cell Viability Assay
4.6. Molecular Docking Analyses
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mazzei, L.; Cianci, M.; Musiani, F.; Ciurli, S. Inactivation of urease by 1,4-benzoquinone: Chemistry at the protein surface. Dalton Trans. 2016, 45, 5455–5459. [Google Scholar] [CrossRef] [PubMed]
- Taha, M.; Ismail, N.H.; Imran, S.; Wadood, A.; Rahim, F.; Riaz, M. Synthesis of potent urease inhibitors based on disulfide scaffold and their molecular docking studies. Bioorg. Med. Chem. 2015, 23, 7211–7218. [Google Scholar] [CrossRef] [PubMed]
- Konieczna, I.; Zarnowiec, P.; Kwinkowski, M.; Kolesinska, B.; Fraczyk, J.; Kaminski, Z.; Kaca, W. Bacterial urease and its role in long-lasting human diseases. Curr. Protein Pept. 2012, 13, 789–806. [Google Scholar] [CrossRef]
- Awllia, J.A.J.; Sara, A.; Wahab, A.-T.; Al-Ghamdi, M.; Rasheed, S.; Huwait, E.; Iqbal Choudhary, M. Discovery of new inhibitors of urease enzyme: A study using STD-NMR spectroscopy. Lett. Drug Des. Discov. 2015, 12, 819–827. [Google Scholar] [CrossRef]
- Kosikowska, P.; Berlicki, Ł. Urease inhibitors as potential drugs for gastric and urinary tract infections: A patent review. Expert Opin. Ther. Pat. 2011, 21, 945–957. [Google Scholar] [CrossRef] [PubMed]
- Follmer, C.J. Ureases as a target for the treatment of gastric and urinary infections. Clin. Pathol. 2010, 63, 424–430. [Google Scholar] [CrossRef] [PubMed]
- Mobley, H.L.; Island, M.D.; Hausinger, R.P. Molecular biology of microbial ureases. Microbiol. Rev. 1995, 59, 451–480. [Google Scholar] [PubMed]
- Hassan, S.T.S.; Žemlička, M. Plant-derived urease inhibitors as alternative chemotherapeutic agents. Arch. Pharm. 2016, 349, 507–522. [Google Scholar] [CrossRef] [PubMed]
- Ibrar, A.; Khan, I.; Abbas, N. Structurally diversified heterocycles and related privileged scaffolds as potential urease inhibitors: A brief overview. Arch. Pharm. 2013, 346, 423–446. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Kawase, M.; Tani, S. Urease inhibitory activity of simple α,β-unsaturated ketones. Life Sci. 2003, 73, 2985–2990. [Google Scholar] [CrossRef]
- Islam, N.U.; Amin, R.; Shahid, M.; Amin, M.; Zaib, S.; Iqbal, J. A multi-target therapeutic potential of Prunus domestica gum stabilized nanoparticles exhibited prospective anticancer, antibacterial, urease-inhibition, anti-inflammatory and analgesic properties. BMC Complement. Altern. Med. 2017, 17, 276. [Google Scholar] [CrossRef] [PubMed]
- Modolo, L.V.; de Souza, A.X.; Horta, L.P.; Araujo, D.P.; de Fátima, Â. An overview on the potential of natural products as ureases inhibitors: A review. J. Adv. Res. 2015, 6, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Saeed, A.; Mahesar, P.A.; Channar, P.A.; Larik, F.A.; Abbas, Q.; Hassan, M.; Raza, H.; Seo, S.Y. Hybrid pharmacophoric approach in the design and synthesis of coumarin linked pyrazolinyl as urease inhibitors, kinetic mechanism and molecular docking. Chem. Biodivers. 2017, 14, e1700035. [Google Scholar] [CrossRef] [PubMed]
- Mazzei, L.; Cianci, M.; Contaldo, U.; Musiani, F.; Ciurli, S. Urease inhibition in the presence of N-(n-Butyl) thiophosphoric triamide, a suicide substrate: Structure and kinetics. Biochemistry 2017, 56, 5391–5404. [Google Scholar] [CrossRef] [PubMed]
- You, Z.; Yu, H.; Zheng, B.; Zhang, C.; Lv, C.; Li, K.; Pan, L. Syntheses, structures, and inhibition studies of Jack bean urease by copper(II) complexes derived from a tridentate hydrazone ligand. Inorg. Chim. Acta 2017, in press. [Google Scholar] [CrossRef]
- Deng, H.H.; Hong, G.L.; Lin, F.L.; Liu, A.L.; Xia, X.H.; Chen, W. Colorimetric detection of urea, urease, and urease inhibitor based on the peroxidase-like activity of gold nanoparticles. Anal. Chim. Acta 2016, 915, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.T.S.; Šudomová, M. The development of urease inhibitors: What opportunities exist for better treatment of helicobacter pylori infection in children? Children 2017, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.T.S.; Berchová, K.; Šudomová, M. Antimicrobial, antiparasitic and anticancer properties of Hibiscus sabdariffa (L.) and its phytochemicals: In vitro and in vivo studies. Ceska Slov. Farm. 2016, 65, 10–14. [Google Scholar] [PubMed]
- Da-Costa-Rocha, I.; Bonnlaender, B.; Sievers, H.; Pischel, I.; Heinrich, M. Hibiscus sabdariffa L.—A phytochemical and pharmacological review. Food Chem. 2014, 165, 424–443. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, A.L.; Lamm, M.G.; Funk, J.L.; Ritenbaugh, C. Hibiscus sabdariffa L. in the treatment of hypertension and hyperlipidemia: A comprehensive review of animal and human studies. Fitoterapia 2013, 85, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.T.S.; Berchová, K.; Majerová, M.; Pokorná, M.; Švajdlenka, E. In vitro synergistic effect of Hibiscus sabdariffa aqueous extract in combination with standard antibiotics against Helicobacter pylori clinical isolates. Pharm. Biol. 2016, 54, 1736–1740. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.T.S.; Švajdlenka, E.; Berchová-Bímová, K. Hibiscus sabdariffa L. and its bioactive constituents exhibit antiviral activity against HSV-2 and anti-enzymatic properties against urease by an ESI-MS based assay. Molecules 2017, 22, 722. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.K.; Rashid, R.; Fatima, N.; Mahmood, S.; Mir, S.; Khan, S.; Jabeen, N.; Murtaza, G. Pharmacological activities of protocatechuic acid. Acta Pol. Pharm. 2015, 72, 643–650. [Google Scholar] [PubMed]
- Semaming, Y.; Pannengpetch, P.; Chattipakorn, S.C.; Chattipakorn, N. Pharmacological properties of protocatechuic Acid and its potential roles as complementary medicine. Evid. Based Complement. Altern. Med. 2015, 2015, 593902. [Google Scholar] [CrossRef] [PubMed]
- Kakkar, S.; Bais, S. A review on protocatechuic acid and its pharmacological potential. ISRN Pharmacol. 2014, 2014, 952943. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Kawase, M.; Tani, S. Alpha-hydroxyketones as inhibitors of urease. Bioorg. Med. Chem. 2004, 12, 501–505. [Google Scholar] [CrossRef] [PubMed]
- Scheurer, M.; Brauch, H.J.; Schmidt, C.K.; Sacher, F.; Zerulla, W.; Barth, T.; Dressel, J.; Erhardt, K.; Locquenghien, K.H.; Pasda, G. Occurrence and fate of nitrification and urease inhibitors in the aquatic environment. Environ. Sci. Process. Impacts 2016, 34, 79–84. [Google Scholar]
- Krajewska, B.; Zaborska, W.; Chudy, M. Multi-step analysis of Hg2+ ion inhibition of jack bean urease. J. Inorg. Biochem. 2004, 98, 1160–1168. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.H.; Chen, J.H.; Chou, F.P.; Wang, C.J. Protocatechuic acid inhibits cancer cell metastasis involving the down-regulation of Ras/Akt/NF-κB pathway and MMP-2 production by targeting RhoB activation. Br. J. Pharmacol. 2011, 162, 237–254. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.H.; Chen, J.H.; Huang, C.C.; Wang, C.J. Apoptotic effect of 3,4-dihydroxybenzoic acid on human gastric carcinoma cells involving JNK/p38 MAPK signaling activation. Int. J. Cancer 2007, 120, 2306–2316. [Google Scholar] [CrossRef] [PubMed]
- Vitaglione, P.; Donnarumma, G.; Napolitano, A.; Galvano, F.; Gallo, A.; Scalfi, L.; Fogliano, V. Protocatechuic acid is the major human metabolite of cyanidin-glucosides. J. Nutr. 2007, 137, 2043–2048. [Google Scholar] [PubMed]
- Chun, O.K.; Chung, S.J.; Song, W.O. Estimated dietary flavonoid intake and major food sources of U.S. adults. J. Nutr. 2007, 137, 1244–1252. [Google Scholar] [PubMed]
- Kay, C.D.; Mazza, G.; Holub, B.J.; Wang, J. Anthocyanin metabolites in human urine and serum. Br. J. Nutr. 2004, 91, 933–942. [Google Scholar] [CrossRef] [PubMed]
- Filiz, E.; Vatansever, R.; Ozyigit, I.I. Molecular docking of Glycine max and Medicago truncatula ureases with urea; bioinformatics approaches. Mol. Biol. Rep. 2016, 43, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Amtul, Z.; Rahman, A.U.; Siddiqui, R.A.; Choudhary, M.I. Chemistry and mechanism of urease inhibition. Curr. Med. Chem. 2002, 9, 1323–1348. [Google Scholar] [CrossRef] [PubMed]
- Farrugia, M.A.; Macomber, L.; Hausinger, R.P. Biosynthesis of the urease metallocenter. J. Biol. Chem. 2013, 288, 3178–3185. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C. Beware of docking! Trends Pharmacol. Sci. 2015, 36, 78–95. [Google Scholar] [CrossRef] [PubMed]
- Shelley, J.C.; Cholleti, A.; Frye, L.L.; Greenwood, J.R.; Timlin, M.R.; Uchimaya, M. Epik: A software program for pKa prediction and protonation state generation for drug-like molecules. J. Comput. Aided Mol. Des. 2007, 21, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Xian, Y.F.; Lin, Z.X.; Mao, Q.Q.; Hu, Z.; Zhao, M.; Che, C.T.; Ip, S.P. Bioassay-guided isolation of neuroprotective compounds from Uncaria rhynchophylla against β-amyloid-induced neurotoxicity. Evid. Based Complement Altern. Med. 2012, 2012, 802625. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Musiani, F.; Arnofi, E.; Casadio, R.; Ciurli, S. Structure-based computational study of the catalytic and inhibition mechanisms of urease. J. Biol. Inorg. Chem. 2001, 6, 300–314. [Google Scholar] [CrossRef] [PubMed]
- Dassault Systèmes BIOVIA. Discovery Studio Modeling Environment; Release 2017; Dessault Systemes: San Diego, CA, USA, 2017. [Google Scholar]
- DeLano, W.L. The PyMOL Molecular Graphics System; DeLano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]
Sample Availability: Samples of all compounds used in the study are available from the authors. |
| Compound | JBU Binding Affinity (kcal/mol) | HPU Binding Affinity (kcal/mol) |
|---|---|---|
| PCA | −5.8 | −5.9 |
| AHA | −4.8 | −4.9 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hassan, S.T.S.; Švajdlenka, E. Biological Evaluation and Molecular Docking of Protocatechuic Acid from Hibiscus sabdariffa L. as a Potent Urease Inhibitor by an ESI-MS Based Method. Molecules 2017, 22, 1696. https://doi.org/10.3390/molecules22101696
Hassan STS, Švajdlenka E. Biological Evaluation and Molecular Docking of Protocatechuic Acid from Hibiscus sabdariffa L. as a Potent Urease Inhibitor by an ESI-MS Based Method. Molecules. 2017; 22(10):1696. https://doi.org/10.3390/molecules22101696
Chicago/Turabian StyleHassan, Sherif T. S., and Emil Švajdlenka. 2017. "Biological Evaluation and Molecular Docking of Protocatechuic Acid from Hibiscus sabdariffa L. as a Potent Urease Inhibitor by an ESI-MS Based Method" Molecules 22, no. 10: 1696. https://doi.org/10.3390/molecules22101696






