Novel PEI/Poly-γ-Gutamic Acid Nanoparticles for High Efficient siRNA and Plasmid DNA Co-Delivery
Abstract
:1. Introduction
2. Results
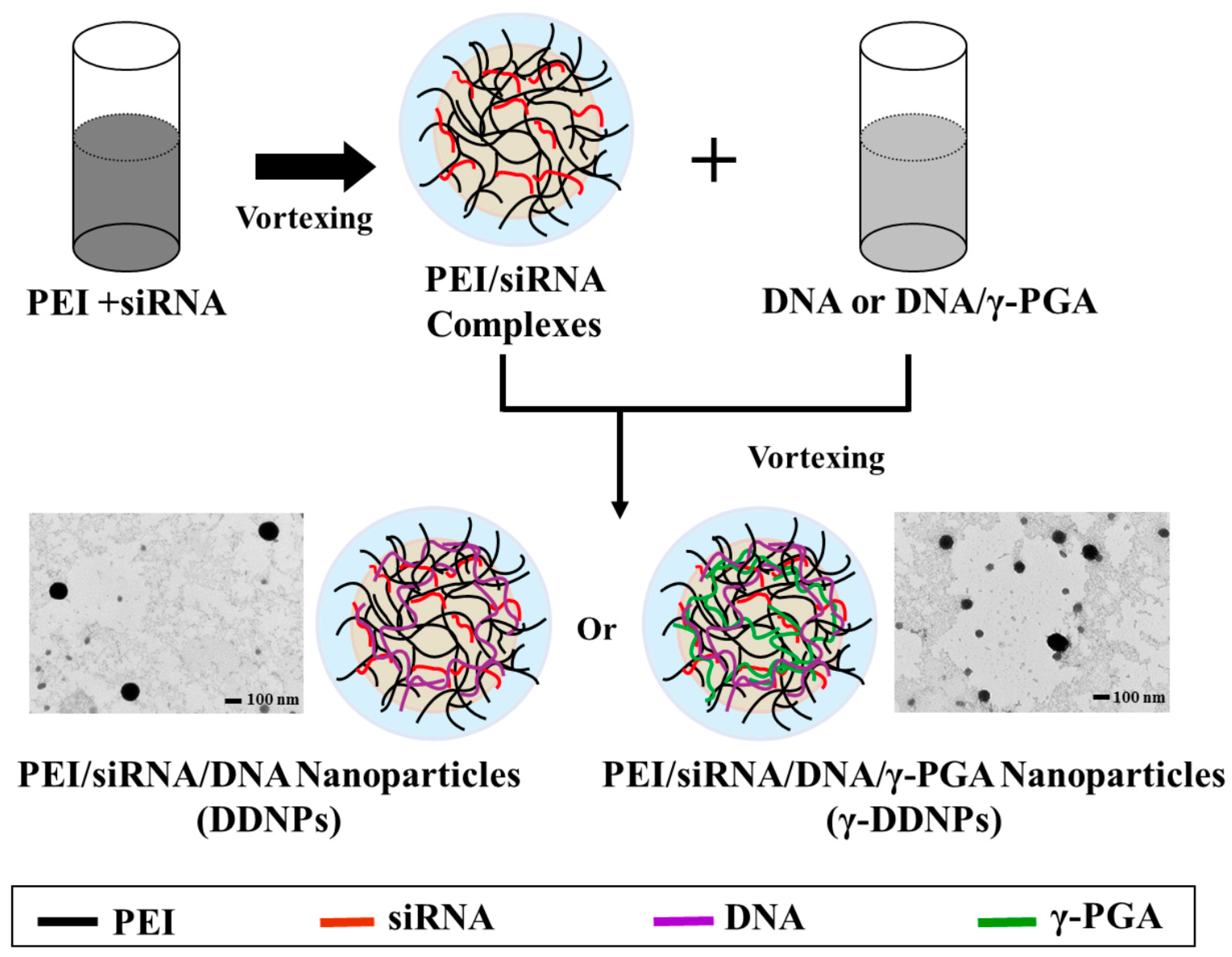
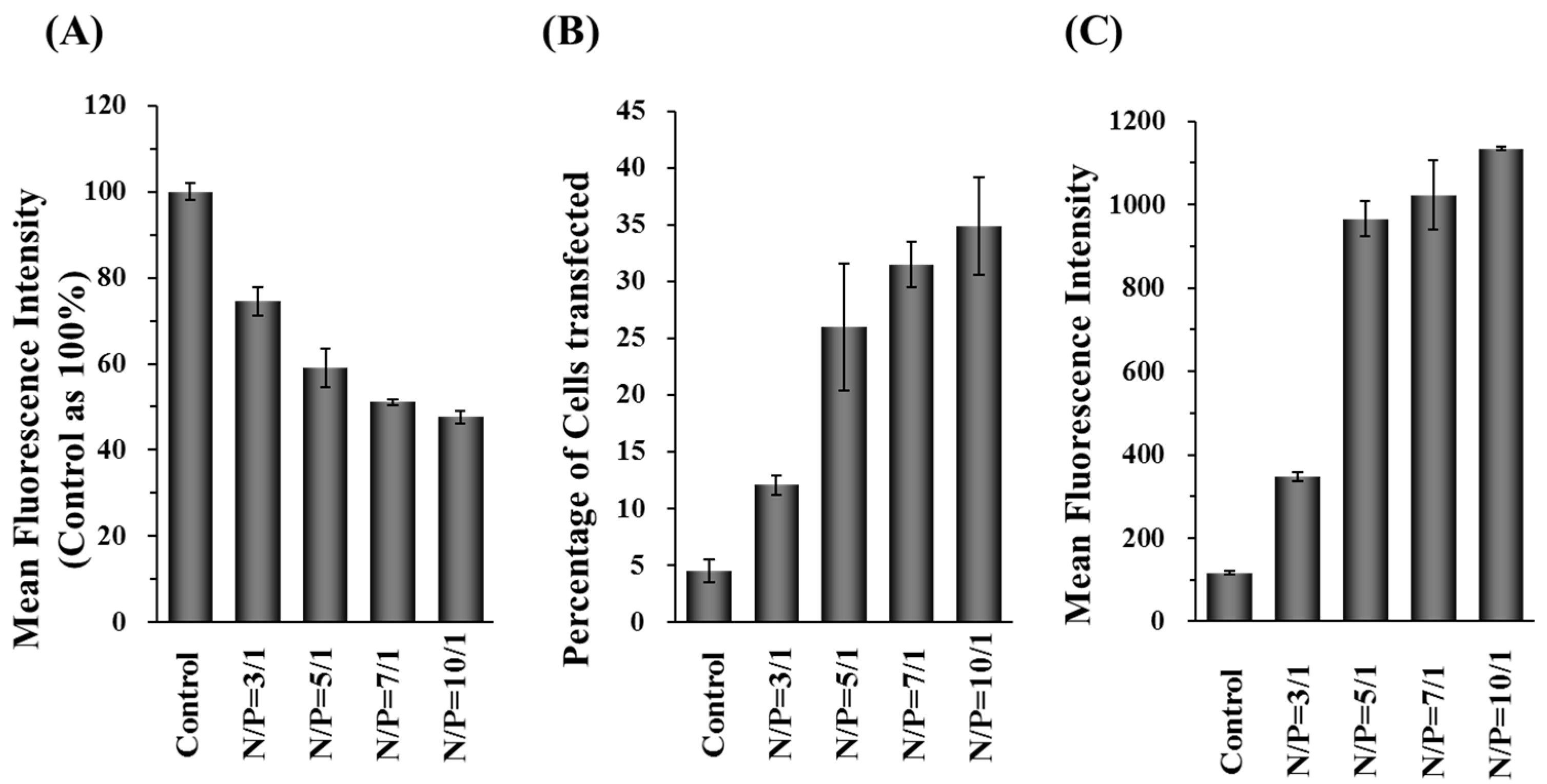
2.1. Characterization, Cellular Uptake, and Transfection Efficiency of DDNPs
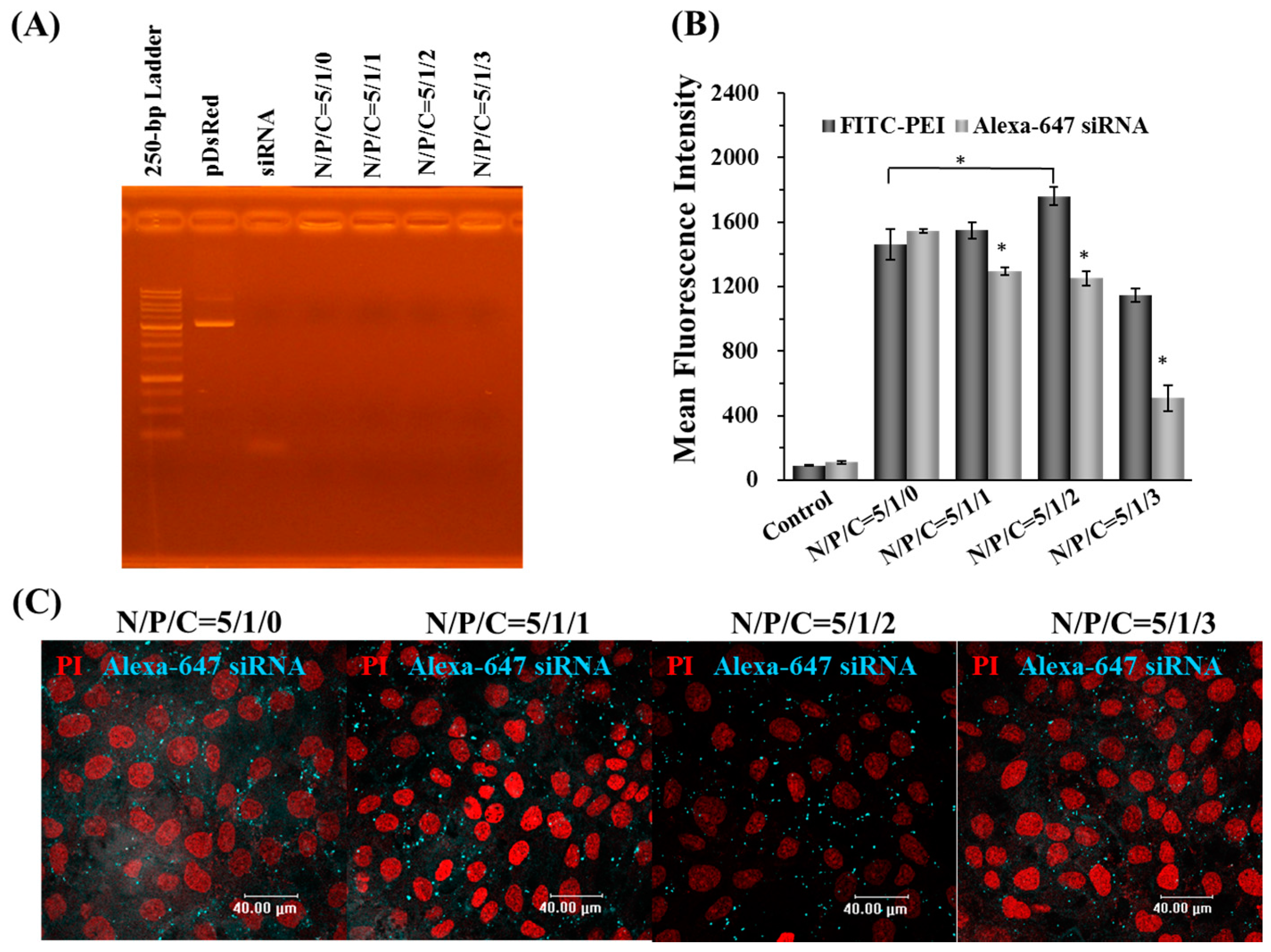
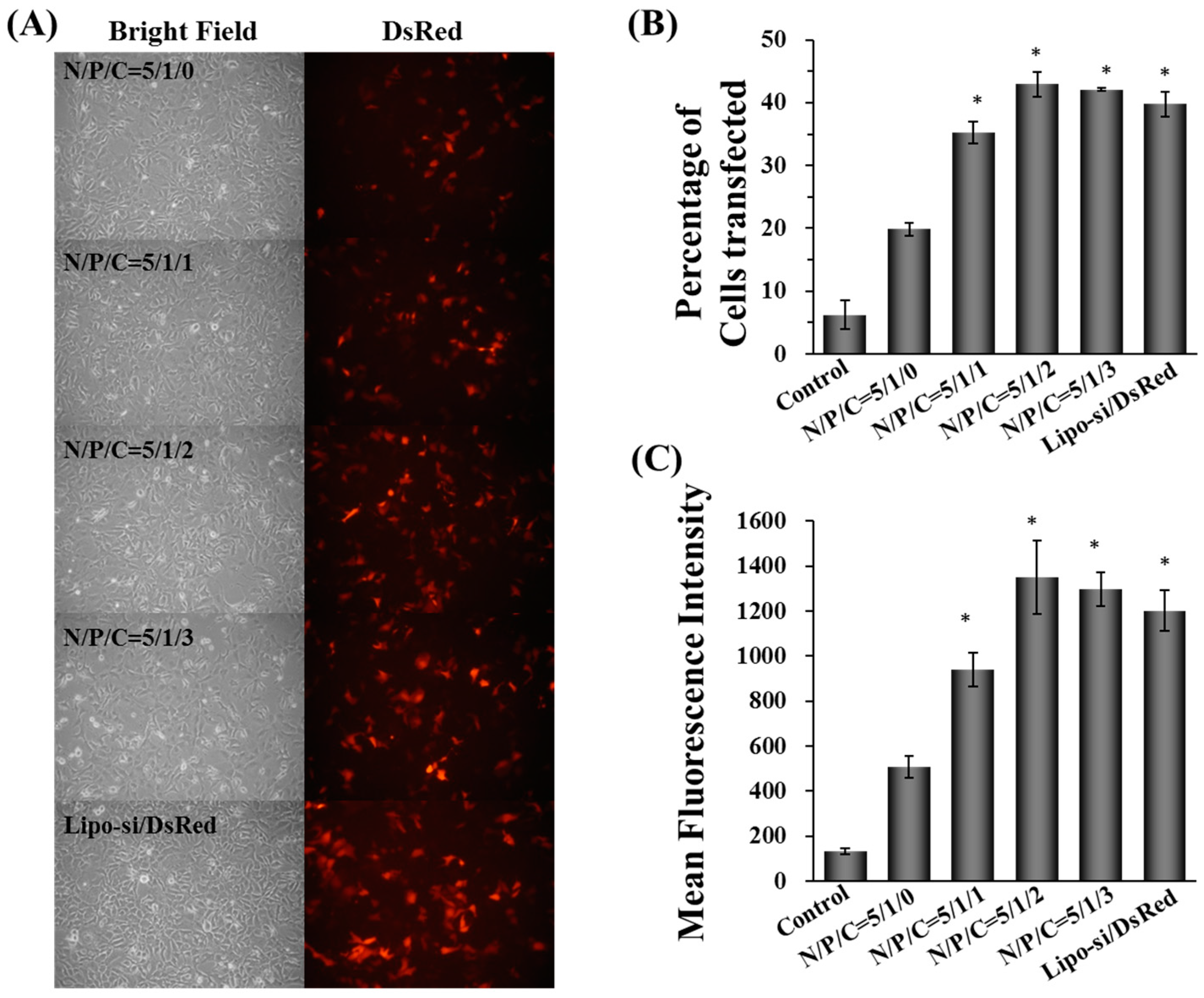
2.2. Characterization, Cellular Uptake, and Transfection Efficiency of Dual-Deliver Nanoparticles Incorporating γ-PGA (γ-DDNPs)
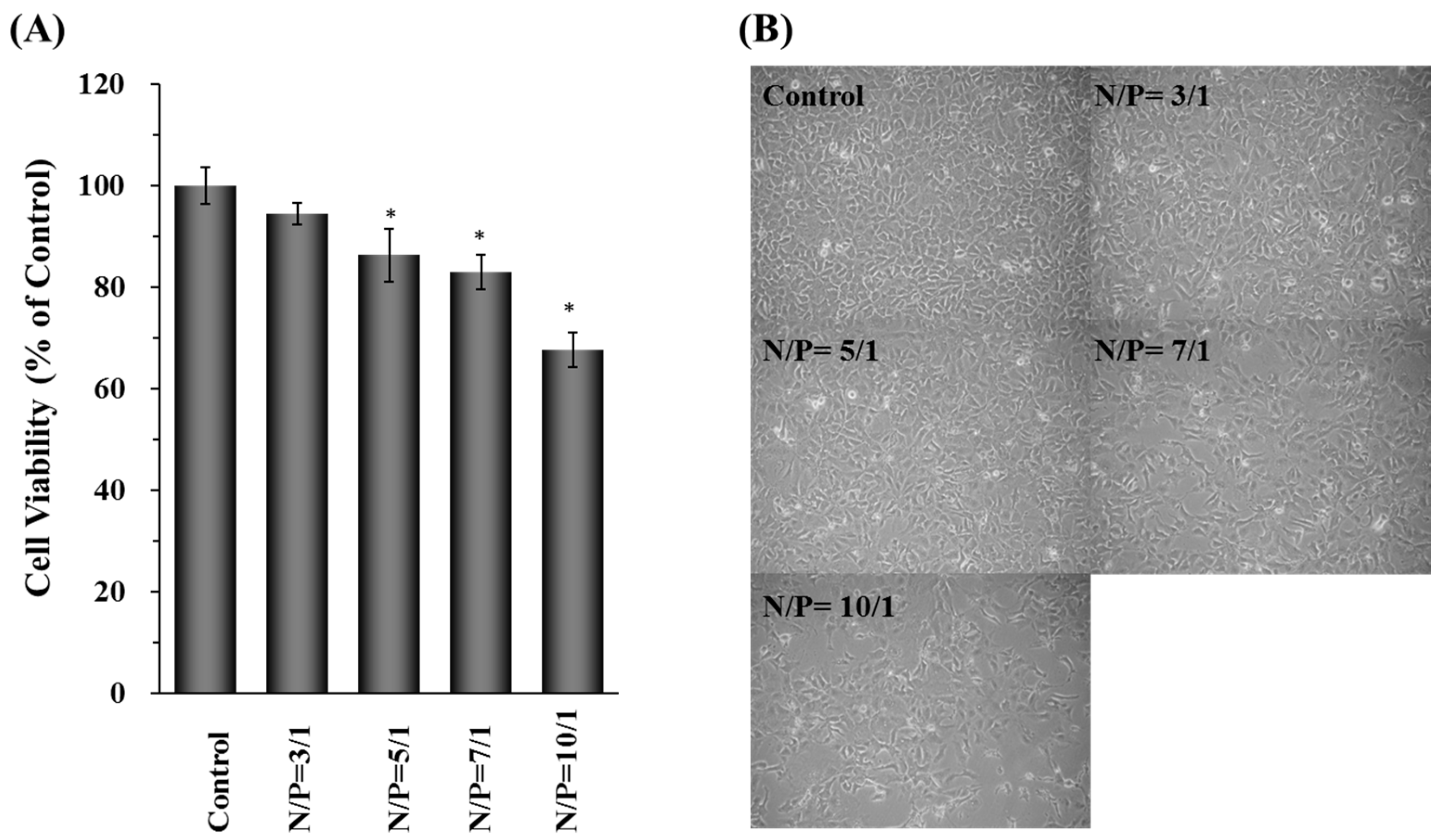
2.3. Cell Cytotoxicity of γ-DDNPs
3. Discussion
4. Experimental Section
4.1. Materials
4.2. Plasmid DNA Preparation
4.3. Preparation of Test Nanoparticles
4.4. Particle Size and Zeta Potential Measurements
4.5. Gel Retardation Assay
4.6. Nanoparticle Morphology Observation
4.7. Cell Culture and In Vitro Transfection
4.8. Fluorescent Nanoparticle Preparation, CLSM Visualization and Flow-Cytometry Analysis
4.9. Percentage and Gene Expression Level of Cells Transfected
4.10. In Vitro Gene Silencing
4.11. Cell Viability Assay
4.12. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pack, D.W.; Hoffman, A.S.; Pun, S.; Stayton, P.S. Design and development of polymers for gene delivery. Nat. Rev. Drug Discov. 2005, 4, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Merdan, T.; Kopecek, J.; Kissel, T. Prospects for cationic polymers in gene and oligonucleotide therapy against cancer. Adv. Drug Deliv. Rev. 2002, 54, 715–758. [Google Scholar] [CrossRef]
- Mulligan, R.C. The basic science of gene therapy. Science 1993, 260, 926–932. [Google Scholar] [CrossRef] [PubMed]
- Pecot, C.V.; Calin, G.A.; Coleman, R.L.; Lopez-Berestein, G.; Sood, A.K. RNA interference in the clinic: Challenges and future directions. Nat. Rev. Cancer 2011, 11, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Van Zundert, B.; Brown, R.H., Jr. Silencing strategies for therapy of SOD1-mediated ALS. Neurosci. Lett. 2017, 636, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, J.; Guo, N.; Zhang, X. Reversal of multidrug resistance in breast cancer MCF-7/ADR cells by h-R3-siMDR1-PAMAM complexes. Int. J. Pharm. 2016, 511, 436–445. [Google Scholar] [CrossRef] [PubMed]
- Meister, G.; Tuschl, T. Mechanisms of gene silencing by double-stranded RNA. Nature 2004, 431, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Rana, T.M. Illuminating the silence: Understanding the structure and function of small RNAs. Nat. Rev. Mol. Cell Biol. 2007, 8, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Park, T.G.; Jeong, J.H.; Kim, S.W. Current status of polymeric gene delivery systems. Adv. Drug Deliv. Rev. 2006, 58, 467–486. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, K.; Harashima, H. Gene delivery systems: Viral vs. non-viral vectors. Adv. Drug Deliv. Rev. 2001, 52, 151. [Google Scholar] [CrossRef]
- Glover, D.J.; Lipps, H.J.; Jans, D.A. Towards safe, non-viral therapeutic gene expression in humans. Nat. Rev. Genet. 2005, 6, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Woods, N.B.; Muessig, A.; Schmidt, M.; Flygare, J.; Olsson, K.; Salmon, P.; Trono, D.; von Kalle, C.; Karlsson, S. Lentiviral vector transduction of NOD/SCID repopulating cells results in multiple vector integrations per transduced cell: Risk of insertional mutagenesis. Blood 2003, 101, 1284–1289. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Garnett, M.C. Gene-delivery systems using cationic polymers. Crit. Rev. Ther. Drug Carrier Syst. 1999, 16, 147–207. [Google Scholar] [CrossRef] [PubMed]
- Kichler, A.; Leborgne, C.; Coeytaux, E.; Danos, O. Polyethylenimine-mediated gene delivery: A mechanistic study. J. Gene Med. 2001, 3, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, B.; Hassan, A.; Benoist, C.; Goula, D.; Behr, J.P.; Demeneix, B.A. A powerful nonviral vector for in vivo gene transfer into the adult mammalian brain: Polyethylenimine. Hum. Gene Ther. 1996, 7, 1947–1954. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, P.; Karashima, T.; Ishikura, H.; Wiehle, S.; Yamashita, M.; Benedict, W.F.; Cristiano, R.J.; Dinney, C.P. Efficient therapeutic gene delivery after systemic administration of a novel polyethylenimine/DNA vector in an orthotopic bladder cancer model. Cancer Res. 2003, 63, 4017–4020. [Google Scholar] [PubMed]
- Boussif, O.; Lezoualc’h, F.; Zanta, M.A.; Mergny, M.D.; Scherman, D.; Demeneix, B.; Behr, J.P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proc. Natl. Acad. Sci. USA 1995, 92, 7297–7301. [Google Scholar] [CrossRef] [PubMed]
- Venkiteswaran, S.T.T.; Thomas, T.J. Selectivity of polyethyleneimines on DNA nanoparticle preparation and gene transport. ChemistrySelect 2016. [Google Scholar] [CrossRef]
- Lungwitz, U.; Breunig, M.; Blunk, T.; Gopferich, A. Polyethylenimine-based non-viral gene delivery systems. Eur. J. Pharm. Biopharm. 2005, 60, 247–266. [Google Scholar] [CrossRef] [PubMed]
- Urban-Klein, B.; Werth, S.; Abuharbeid, S.; Czubayko, F.; Aigner, A. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Ther. 2005, 12, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Zintchenko, A.; Philipp, A.; Dehshahri, A.; Wagner, E. Simple modifications of branched PEI lead to highly efficient siRNA carriers with low toxicity. Bioconjugate Chem. 2008, 19, 1448–1455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, D.; Li, Y.; Ahlemeyer, B.; Krieglstein, J.; Kissel, T. In vitro cytotoxicity testing of polycations: Influence of polymer structure on cell viability and hemolysis. Biomaterials 2003, 24, 1121–1131. [Google Scholar] [CrossRef]
- Godbey, W.T.; Wu, K.K.; Mikos, A.G. Size matters: Molecular weight affects the efficiency of poly(ethylenimine) as a gene delivery vehicle. J. Biomed. Mater. Res. 1999, 45, 268–275. [Google Scholar] [CrossRef]
- He, Y.; Cheng, G.; Xie, L.; Nie, Y.; He, B.; Gu, Z. Polyethyleneimine/DNA polyplexes with reduction-sensitive hyaluronic acid derivatives shielding for targeted gene delivery. Biomaterials 2013, 34, 1235–1245. [Google Scholar] [CrossRef] [PubMed]
- Kurosaki, T.; Kitahara, T.; Fumoto, S.; Nishida, K.; Nakamura, J.; Niidome, T.; Kodama, Y.; Nakagawa, H.; To, H.; Sasaki, H. Ternary complexes of pDNA, polyethylenimine, and gamma-polyglutamic acid for gene delivery systems. Biomaterials 2009, 30, 2846–2853. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, X.; Gou, M.; Qiu, J.; Li, J.; Yu, C.; Zhang, Y.; Zhang, N.; Teng, X.; Chen, Z.; et al. Antitumoral efficacy by systemic delivery of heparin conjugated polyethylenimine-plasmid interleukin-15 complexes in murine models of lung metastasis. Cancer Sci. 2011, 102, 1403–1409. [Google Scholar] [CrossRef] [PubMed]
- Shih, I.L.; Van, Y.T. The production of poly-(gamma-glutamic acid) from microorganisms and its various applications. Bioresour. Technol. 2001, 79, 207–225. [Google Scholar] [CrossRef]
- Peng, S.F.; Yang, M.J.; Su, C.J.; Chen, H.L.; Lee, P.W.; Wei, M.C.; Sung, H.W. Effects of incorporation of poly(gamma-glutamic acid) in chitosan/DNA complex nanoparticles on cellular uptake and transfection efficiency. Biomaterials 2009, 30, 1797–1808. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.F.; Tseng, M.T.; Ho, Y.C.; Wei, M.C.; Liao, Z.X.; Sung, H.W. Mechanisms of cellular uptake and intracellular trafficking with chitosan/DNA/poly(gamma-glutamic acid) complexes as a gene delivery vector. Biomaterials 2011, 32, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Liao, Z.X.; Peng, S.F.; Ho, Y.C.; Mi, F.L.; Maiti, B.; Sung, H.W. Mechanistic study of transfection of chitosan/DNA complexes coated by anionic poly(gamma-glutamic acid). Biomaterials 2012, 33, 3306–3315. [Google Scholar] [CrossRef] [PubMed]
- Liao, Z.X.; Ho, Y.C.; Chen, H.L.; Peng, S.F.; Hsiao, C.W.; Sung, H.W. Enhancement of efficiencies of the cellular uptake and gene silencing of chitosan/siRNA complexes via the inclusion of a negatively charged poly(gamma-glutamic acid). Biomaterials 2010, 31, 8780–8788. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gao, S.; Ye, W.H.; Yoon, H.S.; Yang, Y.Y. Co-delivery of drugs and DNA from cationic core-shell nanoparticles self-assembled from a biodegradable copolymer. Nat. Mater. 2006, 5, 791–796. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.Y.; Bae, Y.H. Self-assembled polyethylenimine-graft-poly(epsilon-caprolactone) micelles as potential dual carriers of genes and anticancer drugs. Biomaterials 2007, 28, 4132–4142. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Morris, V.B.; Labhasetwar, V. Codelivery of DNA and siRNA via arginine-rich PEI-based polyplexes. Mol. Pharm. 2015, 12, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Bishop, C.J.; Tzeng, S.Y.; Green, J.J. Degradable polymer-coated gold nanoparticles for co-delivery of DNA and siRNA. Acta Biomater. 2015, 11, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Shim, M.S.; Kwon, Y.J. Dual mode polyspermine with tunable degradability for plasmid DNA and siRNA delivery. Biomaterials 2011, 32, 4009–4020. [Google Scholar] [CrossRef] [PubMed]
- Felgner, P.L.; Gadek, T.R.; Holm, M.; Roman, R.; Chan, H.W.; Wenz, M.; Northrop, J.P.; Ringold, G.M.; Danielsen, M. Lipofection: A highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. USA 1987, 84, 7413–7417. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.F.; Su, C.J.; Wei, M.C.; Chen, C.Y.; Liao, Z.X.; Lee, P.W.; Chen, H.L.; Sung, H.W. Effects of the nanostructure of dendrimer/DNA complexes on their endocytosis and gene expression. Biomaterials 2010, 31, 5660–5670. [Google Scholar] [CrossRef] [PubMed]
- Malmo, J.; Sorgard, H.; Varum, K.M.; Strand, S.P. siRNA delivery with chitosan nanoparticles: Molecular properties favoring efficient gene silencing. J. Control. Release 2012, 158, 261–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, T.J.; Tajmir-Riahi, H.A.; Thomas, T. Polyamine-DNA interactions and development of gene delivery vehicles. Amino Acids 2016, 48, 2423–2431. [Google Scholar] [CrossRef] [PubMed]
- Vijayanathan, V.; Thomas, T.; Shirahata, A.; Thomas, T.J. DNA condensation by polyamines: A laser light scattering study of structural effects. Biochemistry 2001, 40, 13644–13651. [Google Scholar] [CrossRef] [PubMed]
- Akinc, A.; Thomas, M.; Klibanov, A.M.; Langer, R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J. Gene Med. 2005, 7, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Sonawane, N.D.; Szoka, F.C., Jr.; Verkman, A.S. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. J. Biol. Chem. 2003, 278, 44826–44831. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Lin, L.; Guo, Z.; Xu, C.; Li, Y.; Tian, H.; Tang, Z.; He, C.; Chen, X. N-Isopropylacrylamide Modified Polyethylenimines as Effective siRNA Carriers for Cancer Therapy. J. Nanosci. Nanotechnol. 2016, 16, 5464–5469. [Google Scholar] [CrossRef] [PubMed]
- Das, J.; Das, S.; Paul, A.; Samadder, A.; Bhattacharyya, S.S.; Khuda-Bukhsh, A.R. Assessment of drug delivery and anticancer potentials of nanoparticles-loaded siRNA targeting STAT3 in lung cancer, in vitro and in vivo. Toxicol. Lett. 2014, 225, 454–466. [Google Scholar] [CrossRef] [PubMed]
- Schaffert, D.; Ogris, M. Nucleic acid carrier systems based on polyethylenimine conjugates for the treatment of metastatic tumors. Curr. Med. Chem. 2013, 20, 3456–3470. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Rizzo, M.A.; Bhattacharya, S.; Huang, L. Characterization of cationic lipid-protamine-DNA (LPD) complexes for intravenous gene delivery. Gene Ther. 1998, 5, 930–937. [Google Scholar] [CrossRef] [PubMed]
- Howard, K.A. Delivery of RNA interference therapeutics using polycation-based nanoparticles. Adv. Drug Deliv. Rev. 2009, 61, 710–720. [Google Scholar] [CrossRef] [PubMed]
- Grayson, A.C.; Doody, A.M.; Putnam, D. Biophysical and structural characterization of polyethylenimine-mediated siRNA delivery in vitro. Pharm. Res. 2006, 23, 1868–1876. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhou, L.; Hong, B.; van den Heuvel, A.P.; Prabhu, V.V.; Warfel, N.A.; Kline, C.L.; Dicker, D.T.; Kopelovich, L.; El-Deiry, W.S. Small-Molecule NSC59984 Restores p53 Pathway Signaling and Antitumor Effects against Colorectal Cancer via p73 Activation and Degradation of Mutant p53. Cancer Res. 2015, 75, 3842–3852. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Feijen, J.; Lok, M.C.; Hennink, W.E.; Christensen, L.V.; Yockman, J.W.; Kim, Y.H.; Kim, S.W. Low molecular weight linear polyethylenimine-b-poly(ethylene glycol)-b-polyethylenimine triblock copolymers: Synthesis, characterization, and in vitro gene transfer properties. Biomacromolecules 2005, 6, 3440–3448. [Google Scholar] [CrossRef] [PubMed]
- Kodama, Y.; Shiokawa, Y.; Nakamura, T.; Kurosaki, T.; Aki, K.; Nakagawa, H.; Muro, T.; Kitahara, T.; Higuchi, N.; Sasaki, H. Novel siRNA delivery system using a ternary polymer complex with strong silencing effect and no cytotoxicity. Biol. Pharm. Bull. 2014, 37, 1274–1281. [Google Scholar] [CrossRef] [PubMed]
- Rejman, J.; Bragonzi, A.; Conese, M. Role of clathrin- and caveolae-mediated endocytosis in gene transfer mediated by lipo- and polyplexes. Mol. Ther. 2005, 12, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Gabrielson, N.P.; Pack, D.W. Efficient polyethylenimine-mediated gene delivery proceeds via a caveolar pathway in HeLa cells. J. Control. Release 2009, 136, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Hwang, M.E.; Keswani, R.K.; Pack, D.W. Dependence of PEI and PAMAM Gene Delivery on Clathrin- and Caveolin-Dependent Trafficking Pathways. Pharm. Res. 2015, 32, 2051–2059. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, J.; Szoka, F.C. Nucleic acid delivery: The missing pieces of the puzzle? Acc. Chem. Res. 2012, 45, 1153–1162. [Google Scholar] [CrossRef] [PubMed]
- Vijayanathan, V.; Thomas, T.; Thomas, T.J. DNA nanoparticles and development of DNA delivery vehicles for gene therapy. Biochemistry 2002, 41, 14085–14094. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Huang, G.; Cong, Y.; Tong, G.; Lin, Z.; Yin, Y.; Zhang, C. The preparation and characterization of micelles from poly(gamma-glutamic acid)-graft-poly(l-lactide) and the cellular uptake thereof. J. Mater. Sci. Mater. Med. 2015, 26, 187. [Google Scholar] [CrossRef] [PubMed]
- Koeda, S.; Ichiki, K.; Iwanaga, N.; Mizuno, K.; Shibata, M.; Obata, A.; Kasuga, T.; Mizuno, T. Construction and Characterization of Protein-Encapsulated Electrospun Fibermats Prepared from a Silica/Poly(gamma-glutamate) Hybrid. Langmuir 2016, 32, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Maya, S.; Sarmento, B.; Lakshmanan, V.K.; Menon, D.; Jayakumar, R. Actively targeted cetuximab conjugated gamma-poly(glutamic acid)-docetaxel nanomedicines for epidermal growth factor receptor over expressing colon cancer cells. J. Biomed. Nanotechnol. 2014, 10, 1416–1428. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.L.; Ho, C.T.; Chung, J.G.; Raghu, R.; Lo, Y.C.; Sheen, L.Y. Allicin induces anti-human liver cancer cells through the p53 gene modulating apoptosis and autophagy. J. Agric. Food Chem. 2013, 61, 9839–9848. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds (PEI and γ-PGA) are not available from the authors.



| N/P Ratio | Size (nm) | PDI | Zeta Potential (mV) |
|---|---|---|---|
| 3/1 | 178.6 ± 19.8 | 0.44 ± 0.02 | 39.9 ± 5.8 |
| 5/1 | 197.0 ± 9.2 | 0.43 ± 0.03 | 43.6 ± 0.5 |
| 7/1 | 204.7 ± 3.5 | 0.43 ± 0.04 | 45.7 ± 0.6 |
| 10/1 | 203.4 ± 3.8 | 0.44 ± 0.02 | 47.8 ± 2.2 |
| N/P/C Ratio | Size (nm) | PDI | Zeta Potential (mV) |
|---|---|---|---|
| 5/1/0 | 197.0 ± 9.2 | 0.43 ± 0.03 | 43.6 ± 0.5 |
| 5/1/1 | 165.4 ± 2.3 | 0.38 ± 0.02 | 36.1 ± 7.3 |
| 5/1/2 | 180.3 ± 8.2 | 0.26 ± 0.03 | 40.0 ± 1.4 |
| 5/1/3 | 201.5 ± 25.9 | 0.15 ± 0.02 | 36.6 ± 1.4 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, S.-F.; Hsu, H.-K.; Lin, C.-C.; Cheng, Y.-M.; Hsu, K.-H. Novel PEI/Poly-γ-Gutamic Acid Nanoparticles for High Efficient siRNA and Plasmid DNA Co-Delivery. Molecules 2017, 22, 86. https://doi.org/10.3390/molecules22010086
Peng S-F, Hsu H-K, Lin C-C, Cheng Y-M, Hsu K-H. Novel PEI/Poly-γ-Gutamic Acid Nanoparticles for High Efficient siRNA and Plasmid DNA Co-Delivery. Molecules. 2017; 22(1):86. https://doi.org/10.3390/molecules22010086
Chicago/Turabian StylePeng, Shu-Fen, Hung-Kun Hsu, Chun-Cheng Lin, Ya-Ming Cheng, and Kuang-Hsing Hsu. 2017. "Novel PEI/Poly-γ-Gutamic Acid Nanoparticles for High Efficient siRNA and Plasmid DNA Co-Delivery" Molecules 22, no. 1: 86. https://doi.org/10.3390/molecules22010086





