Gold Nanocluster Decorated Polypeptide/DNA Complexes for NIR Light and Redox Dual-Responsive Gene Transfection
Abstract
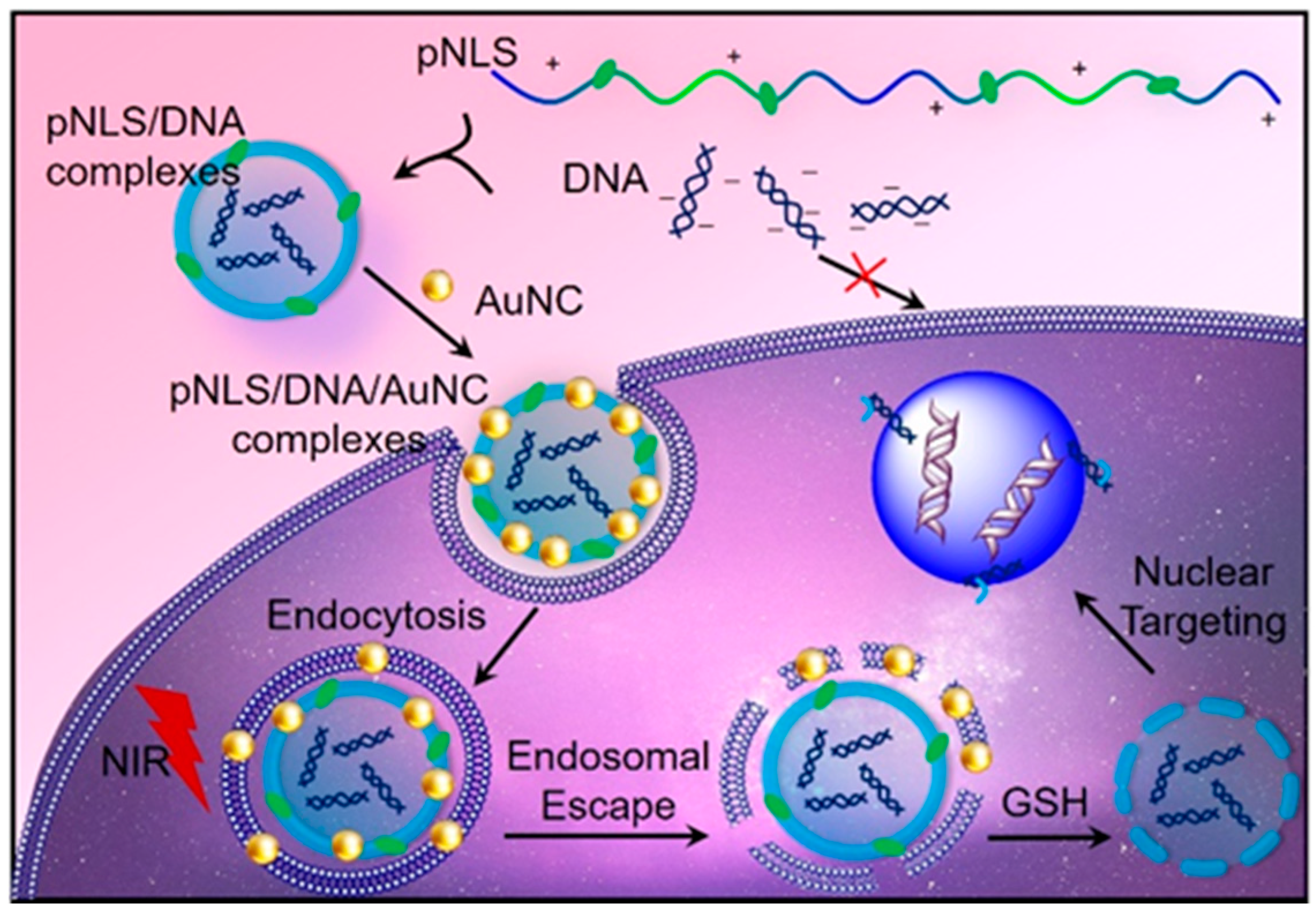
:1. Introduction
2. Results and Discussion
2.1. Synthesis of pNLS
2.2. DNA Compaction Capability
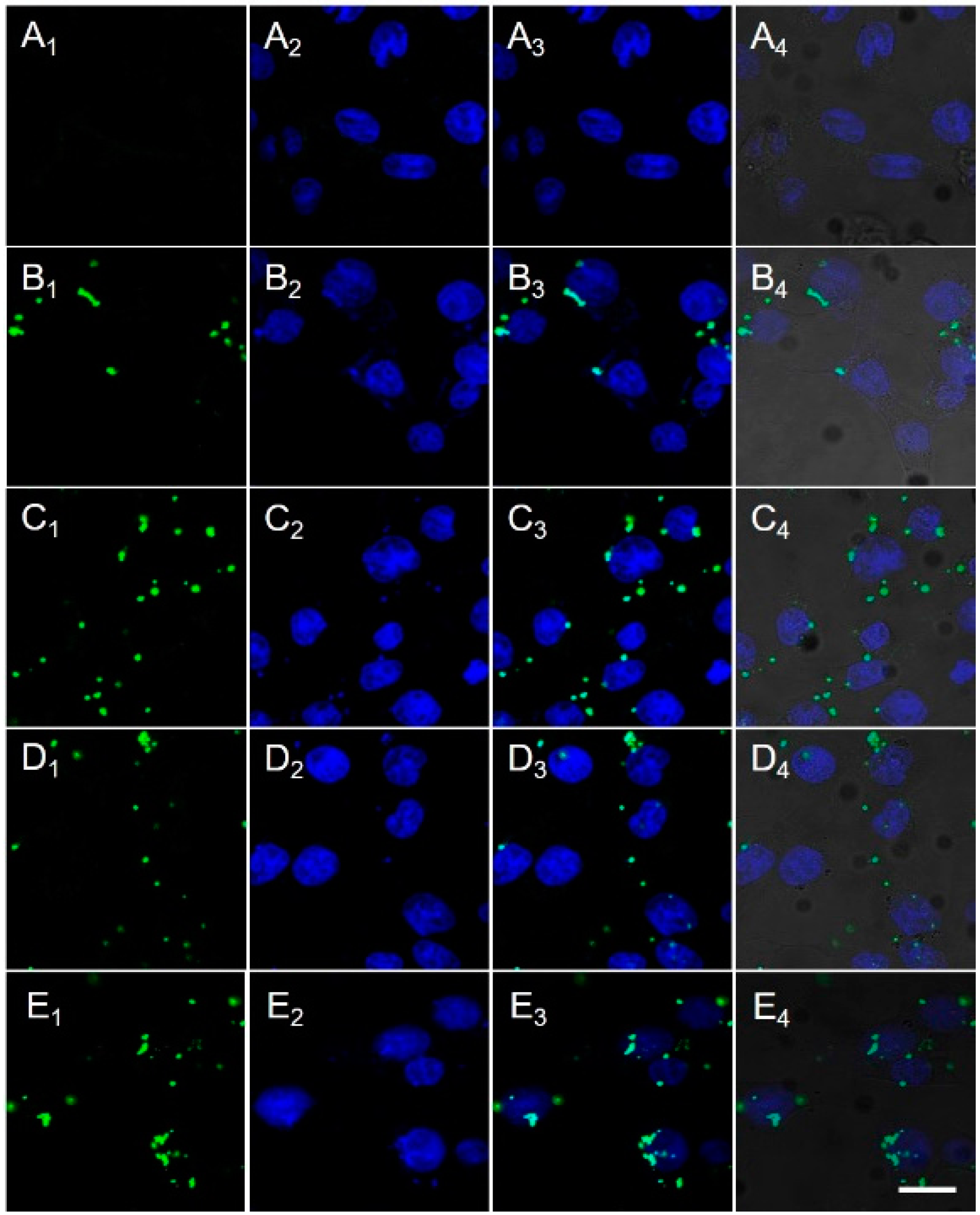
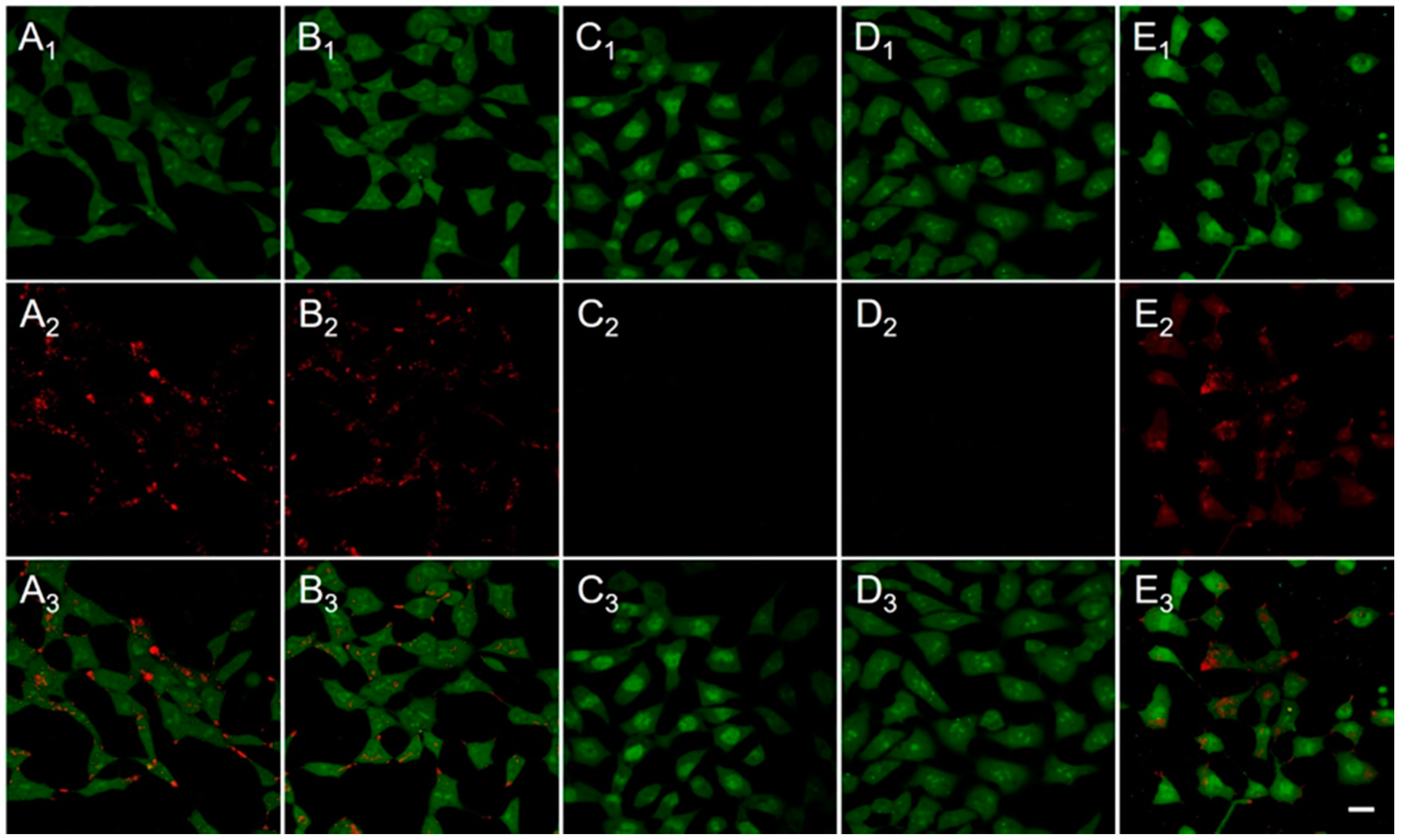
2.3. Cellular Uptake and Nuclear Translocation
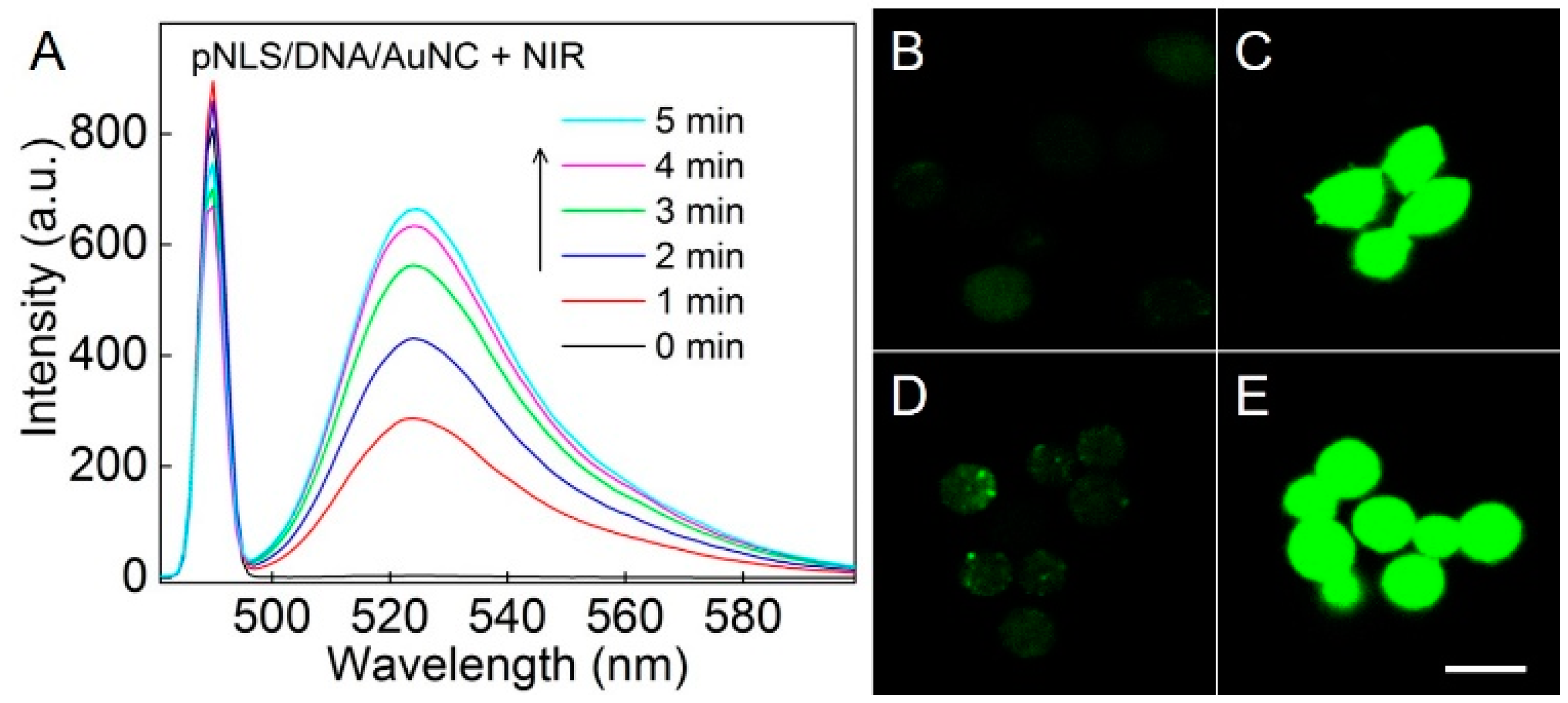
2.4. NIR-Responsive ROS Generation and Endo/Lysosomal Damage
2.5. Cytotoxicity
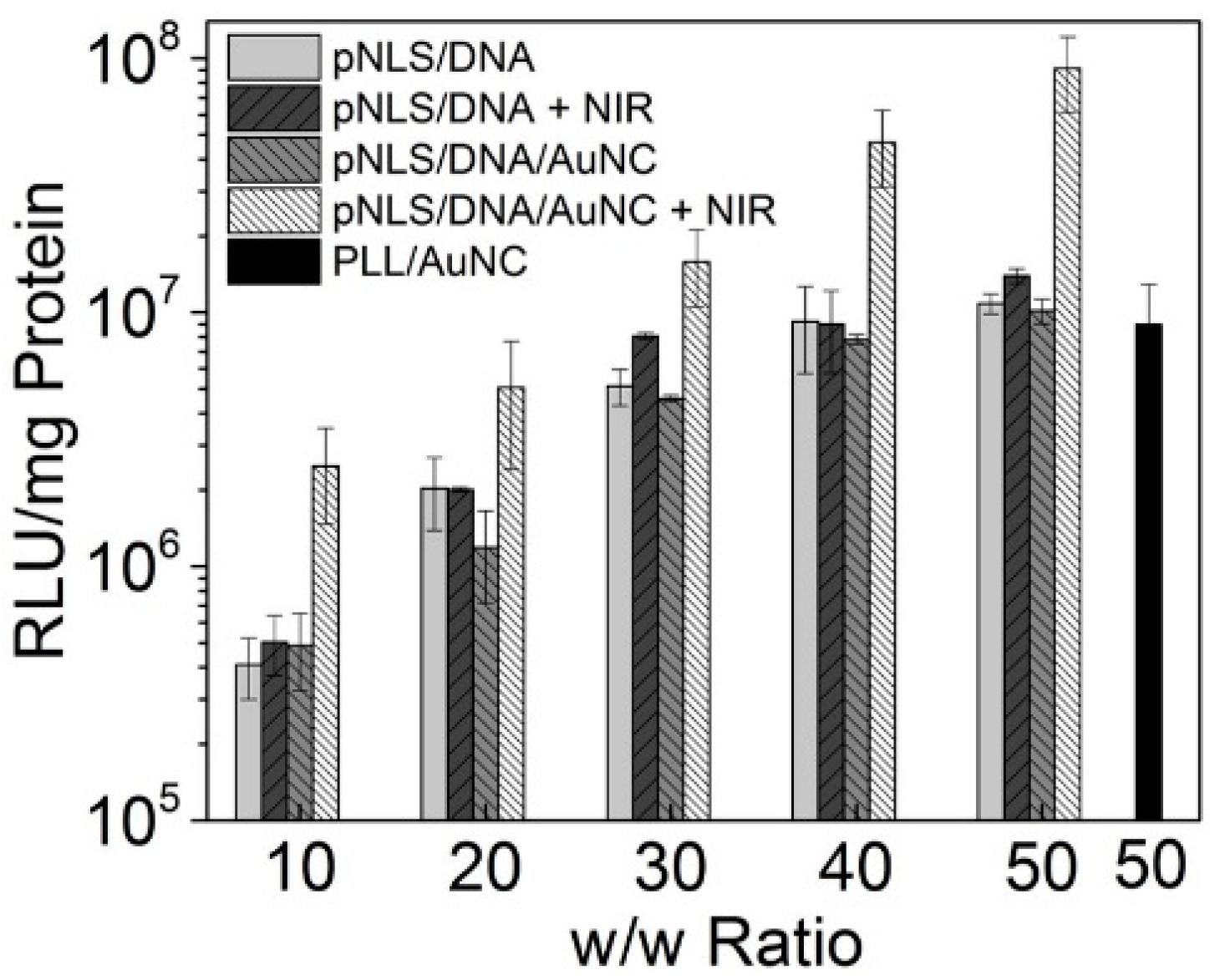
2.6. Gene Transfection
3. Materials and Methods
3.1. Materials
3.2. Synthesis of Peptide Sequences
3.3. Synthesis and Characterizations of Disulfide Linked pNLS
3.4. Synthesis of AuNC
3.5. Cell Culture and Amplification of Plasmid DNA
3.6. Preparation of pNLS/DNA Complexes
3.7. Agarose Gel Retardation Assay
3.8. Particle Size and Zeta Potential Measurements.
3.9. Morphology
3.10. Cellular Uptake Assays
3.11. ROS Detection
3.12. Disruption of Endo/Lysosome
3.13. Cytotoxicity Assays
3.14. Transfection Assays
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kay, M.A. State-of-the-art gene-based therapies: The road ahead. Nat. Rev. Genet. 2011, 12, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Naldini, L. Gene therapy returns to centre stage. Nature 2015, 526, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Zorko, M.; Langel, U. Cell-penetrating peptides: Mechanism and kinetics of cargo delivery. Adv. Drug Deliv. Rev. 2005, 57, 529–545. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Satterlee, A.; Huang, L. In Vivo Gene Delivery by Nonviral Vectors: Overcoming Hurdles&quest. Mol. Ther. 2012, 20, 1298–1304. [Google Scholar] [PubMed]
- Kim, J.; Kim, J.; Jeong, C.; Kim, W.J. Synergistic nanomedicine by combined gene and photothermal therapy. Adv. Drug Deliv. Rev. 2016, 98, 99–112. [Google Scholar] [CrossRef] [PubMed]
- Kotterman, M.A.; Chalberg, T.W.; Schaffer, D.V. Viral vectors for gene therapy: Translational and clinical outlook. Annu. Rev. Biomed. Eng. 2015, 17, 63–89. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, Y.; Zhang, X.; Zhang, W.; Guo, S.; Jin, F. Recent progress of cell-penetrating peptides as new carriers for intracellular cargo delivery. J. Control. Release 2014, 174, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Gao, N.; Wang, Y.; Yi, H.; Fang, S.; Ma, Y.; Cai, L. Self-assembled cationic micelles based on PEG-PLL-PLLeu hybrid polypeptides as highly effective gene vectors. Biomacromolecules 2012, 13, 3795–3804. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.; Lee, D.; Gujrati, V.; Rejinold, N.S.; Lekshmi, K.M.; Uthaman, S.; Jeong, C.; Park, I.K.; Jon, S.; Kim, Y.C. Bioreducible branched poly(modified nona-arginine) cell-penetrating peptide as a novel gene delivery platform. J. Control. Release 2016. [Google Scholar] [CrossRef] [PubMed]
- Qu, W.; Qin, S.Y.; Kuang, Y.; Zhuo, R.X.; Zhang, X.Z. Peptide-based vectors mediated by avidin-biotin interaction for tumor targeted gene delivery. J. Mater. Chem. B 2013, 1, 2147–2154. [Google Scholar] [CrossRef]
- Yang, J.; Lei, Q.; Han, K.; Gong, Y.H.; Chen, S.; Cheng, H.; Cheng, S.X.; Zhuo, R.X.; Zhang, X.Z. Reduction-sensitive polypeptides incorporated with nuclear localization signal sequences for enhanced gene delivery. J. Mater. Chem. 2012, 22, 13591–13599. [Google Scholar] [CrossRef]
- Lei, Q.; Sun, Y.X.; Chen, S.; Qin, S.Y.; Jia, H.Z.; Zhuo, R.X.; Zhang, X.Z. Fabrication of Novel Reduction-Sensitive Gene Vectors Based on Three-Armed Peptides. Macromol. Biosci. 2014, 14, 546–556. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Lei, Q.; Li, S.Y.; Qin, S.Y.; Jia, H.Z.; Cheng, Y.J.; Zhang, X.Z. Fabrication of dual responsive co-delivery system based on three-armed peptides for tumor therapy. Biomaterials 2016, 92, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Han, K.; Yang, J.; Lei, Q.; Zhuo, R.X.; Zhang, X.Z. Bioreducible polypeptide containing cell-penetrating sequence for efficient gene delivery. Pharm. Res. 2013, 30, 1968–1978. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wang, H.Y.; Yi, W.J.; Gong, Y.H.; Zhou, X.; Zhuo, R.X.; Zhang, X.Z. PEGylated peptide based reductive polycations as efficient nonviral gene vectors. Adv. Healthc. Mater. 2013, 2, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Read, M.L.; Bremner, K.H.; Oupick, D.; Green, N.K.; Searle, P.F.; Seymour, L.W. Vectors based on reducible polycations facilitate intracellular release of nucleic acids. J. Gene Med. 2003, 5, 232–245. [Google Scholar] [CrossRef] [PubMed]
- Lei, Q.; Jia, H.Z.; Chen, W.H.; Rong, L.; Chen, S.; Luo, G.F.; Qiu, W.X.; Zhang, X.Z. A Facile Multifunctionalized Gene Delivery Platform Based on α, β Cyclodextrin Dimers. ACS Biomater. Sci. Eng. 2015, 1, 1151–1162. [Google Scholar] [CrossRef]
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkina, J.R.; Anderson, D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Remaut, K.; Oorschot, V.; Braeckmans, K.; Klumperman, J.; Smedt, S.C.D. Lysosomal capturing of cytoplasmic injected nanoparticles by autophagy: An additional barrier to non viral gene delivery. J. Control. Release 2014, 195, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Høgset, A.; Prasmickaite, L.; Selbo, P.K.; Hellum, M.; Engesæter, B.Ø.; Bonsted, A.; Berg, K. Photochemical internalisation in drug and gene delivery. Adv. Drug Deliv. Rev. 2004, 56, 95–115. [Google Scholar] [CrossRef] [PubMed]
- Berg, K.; Folini, M.; Prasmickaite, L.; Selbo, P.K.; Bonsted, A.; Engesæter, B.O.; Zaffaroni, N.; Weyergang, A.; Dietzea, A.; Maelandsmo, G.M.; et al. Photochemical internalization: A new tool for drug delivery. Curr. Pharm. Biotechnol. 2007, 8, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Norum, O.J.; Selbo, P.K.; Weyergang, A.; Giercksky, K.E.; Berg, K. Photochemical internalization (PCI) in cancer therapy: From bench towards bedside medicine. J. Photochem. Photobiol. B 2009, 96, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Berg, K.; Nordstrand, S.; Selbo, P.K.; Tran, D.T.T.; Angell-Petersen, E.; Høgset, A. Disulfonated tetraphenyl chlorin (TPCS 2a), a novel photosensitizer developed for clinical utilization of photochemical internalization. Photochem. Photobiol. Sci. 2011, 10, 1637–1651. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Lei, Q.; Jia, H.Z.; Wang, S.B.; Yin, W.N.; Chen, W.H.; Chen, S.X.; Zhang, X.Z. A Tumor Targeted Chimeric Peptide for Synergistic Endosomal Escape and Therapy by Dual-Stage Light Manipulation. Adv. Funct. Mater. 2015, 25, 1248–1257. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhang, C.J.; Liu, B. A Photoactivatable AIE Polymer for Light-Controlled Gene Delivery: Concurrent Endo/Lysosomal Escape and DNA Unpacking. Angew. Chem. Int. Ed. 2015, 54, 11419–11423. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, N.; Iriyama, A.; Jang, W.D.; Miyata, K.; Itaka, K.; Inoue, Y.; Takahashi, H.; Yanagi, Y.; Tamaki, Y.; Koyama, H.; et al. Light-induced gene transfer from packaged DNA enveloped in a dendrimeric photosensitizer. Nat. Mater. 2005, 4, 934–941. [Google Scholar] [CrossRef] [PubMed]
- Nomoto, T.; Fukushima, S.; Kumagai, M.; Machitani, K.; Arnida; Matsumoto, Y.; Oba, M.; Miyata, K.; Osada, K.; Nishiyama, N.; et al. Three-layered polyplex micelle as a multifunctional nanocarrier platform for light-induced systemic gene transfer. Nat. Commun. 2014, 5, 3545. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Zhang, Z.; Li, J. Highly Efficient Multifunctional Supramolecular Gene Carrier System Self-Assembled from Redox-Sensitive and Zwitterionic Polymer Blocks. Adv. Funct. Mater. 2014, 24, 3874–3884. [Google Scholar] [CrossRef]
- Lv, R.; Yang, P.; He, F.; Gai, S.; Yang, G.; Dai, Y.; Hou, Z.; Lin, J. An imaging-guided platform for synergistic photodynamic/photothermal/chemo- therapy with pH/temperature-responsive drug release. Biomaterials 2015, 63, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Jin, R. Water-soluble Au25(Capt)18 nanoclusters: Synthesis, thermal stability, and optical properties. Nanoscale 2012, 4, 4222–4227. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, H.; Kumar, S.; Li, G.; Zeng, C.; Kauffman, D.R.; Yoshimoto, J.; Iwasaki, Y.; Jin, R.C. Generation of singlet oxygen by photoexcited Au25(SR)18 clusters. Chem. Mater. 2014, 26, 2777–2788. [Google Scholar] [CrossRef]
- Yap, F.L.; Thoniyot, P.; Krishnan, S.; Krishnamoorthy, S. Nanoparticle cluster arrays for high-performance SERS through directed self-assembly on flat substrates and on optical fibers. ACS Nano 2012, 6, 2056–2070. [Google Scholar] [CrossRef] [PubMed]
- Banchelli, M.; Nappini, S.; Montis, C.; Bonini, M.; Canton, P.; Berti, D.; Baglioni, P. Magnetic nanoparticle clusters as actuators of ssDNA release. Phys. Chem. Chem. Phys. 2014, 16, 10023–10031. [Google Scholar] [CrossRef] [PubMed]
- Qiu, P.; Jensen, C.; Charity, N.; Towner, R.; Mao, C. Oil phase evaporation-induced self-assembly of hydrophobic nanoparticles into spherical clusters with controlled surface chemistry in an oil-in-water dispersion and comparison of behaviors of individual and clustered iron oxide nanoparticles. J. Am. Chem. Soc. 2010, 132, 17724–17732. [Google Scholar] [CrossRef] [PubMed]
- Rejman, J.; Oberle, V.; Zuhorn, I.S.; Hoekstra, D. Size-dependent internalization of particles via the pathways of clathrin- and caveolae-mediated endocytosis. Biochem. J. 2004, 377, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Not available.



© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lei, Q.; Hu, J.-J.; Rong, L.; Cheng, H.; Sun, Y.-X.; Zhang, X.-Z. Gold Nanocluster Decorated Polypeptide/DNA Complexes for NIR Light and Redox Dual-Responsive Gene Transfection. Molecules 2016, 21, 1103. https://doi.org/10.3390/molecules21081103
Lei Q, Hu J-J, Rong L, Cheng H, Sun Y-X, Zhang X-Z. Gold Nanocluster Decorated Polypeptide/DNA Complexes for NIR Light and Redox Dual-Responsive Gene Transfection. Molecules. 2016; 21(8):1103. https://doi.org/10.3390/molecules21081103
Chicago/Turabian StyleLei, Qi, Jing-Jing Hu, Lei Rong, Han Cheng, Yun-Xia Sun, and Xian-Zheng Zhang. 2016. "Gold Nanocluster Decorated Polypeptide/DNA Complexes for NIR Light and Redox Dual-Responsive Gene Transfection" Molecules 21, no. 8: 1103. https://doi.org/10.3390/molecules21081103




