A Metagenomic Advance for the Cloning and Characterization of a Cellulase from Red Rice Crop Residues
Abstract
:1. Introduction
2. Results
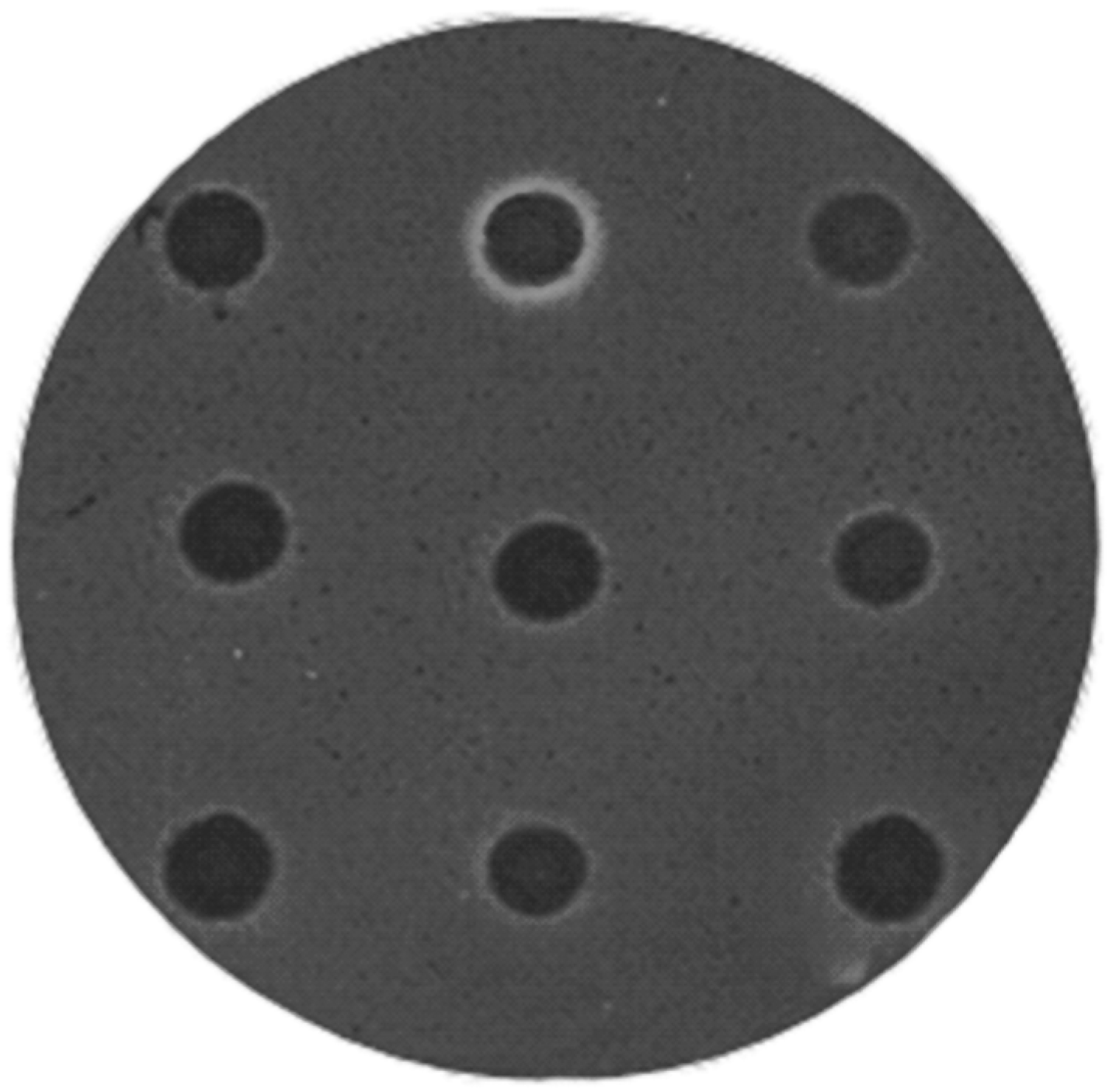
2.1. Construction and Screening of Metagenomic Libraries
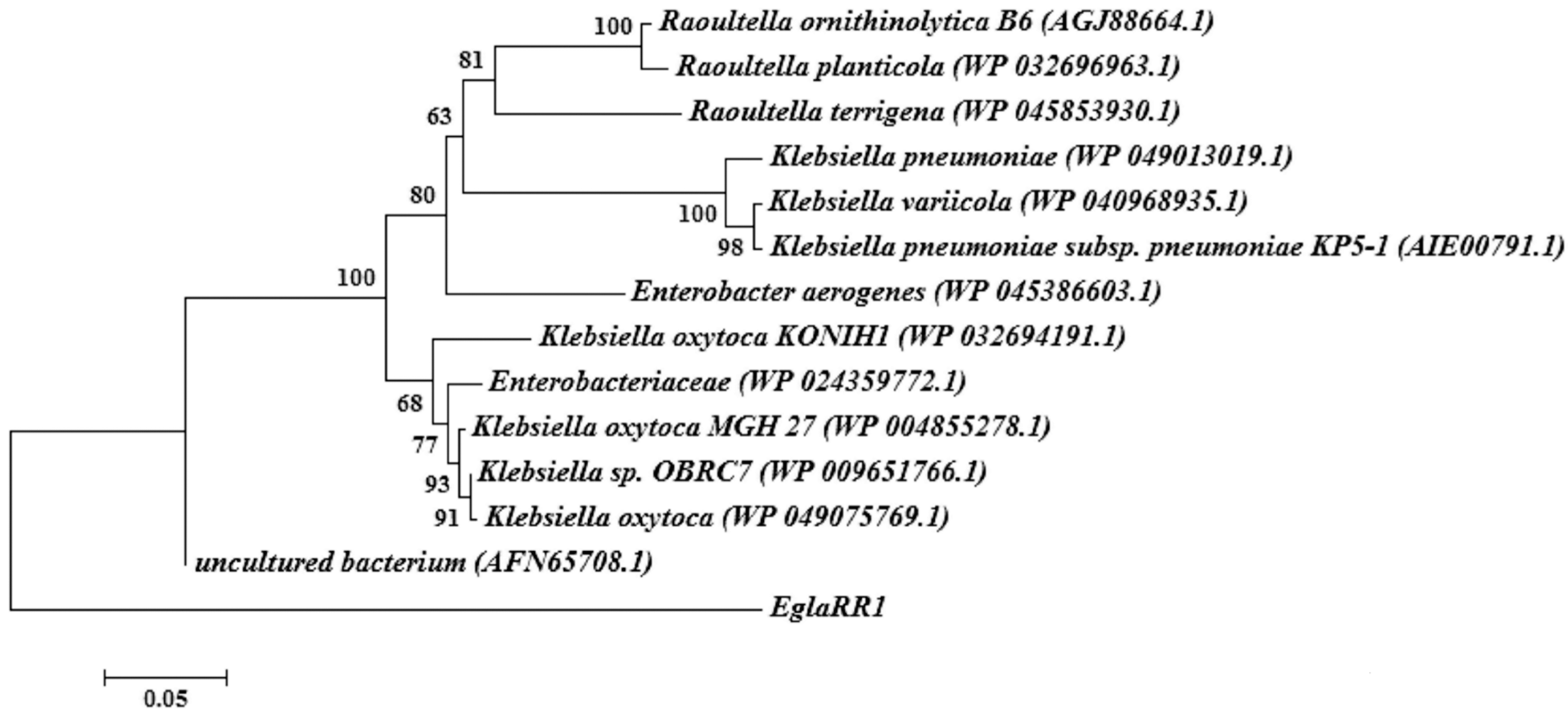
2.2. Sequence Analyses of Cloned Cellulase Genes
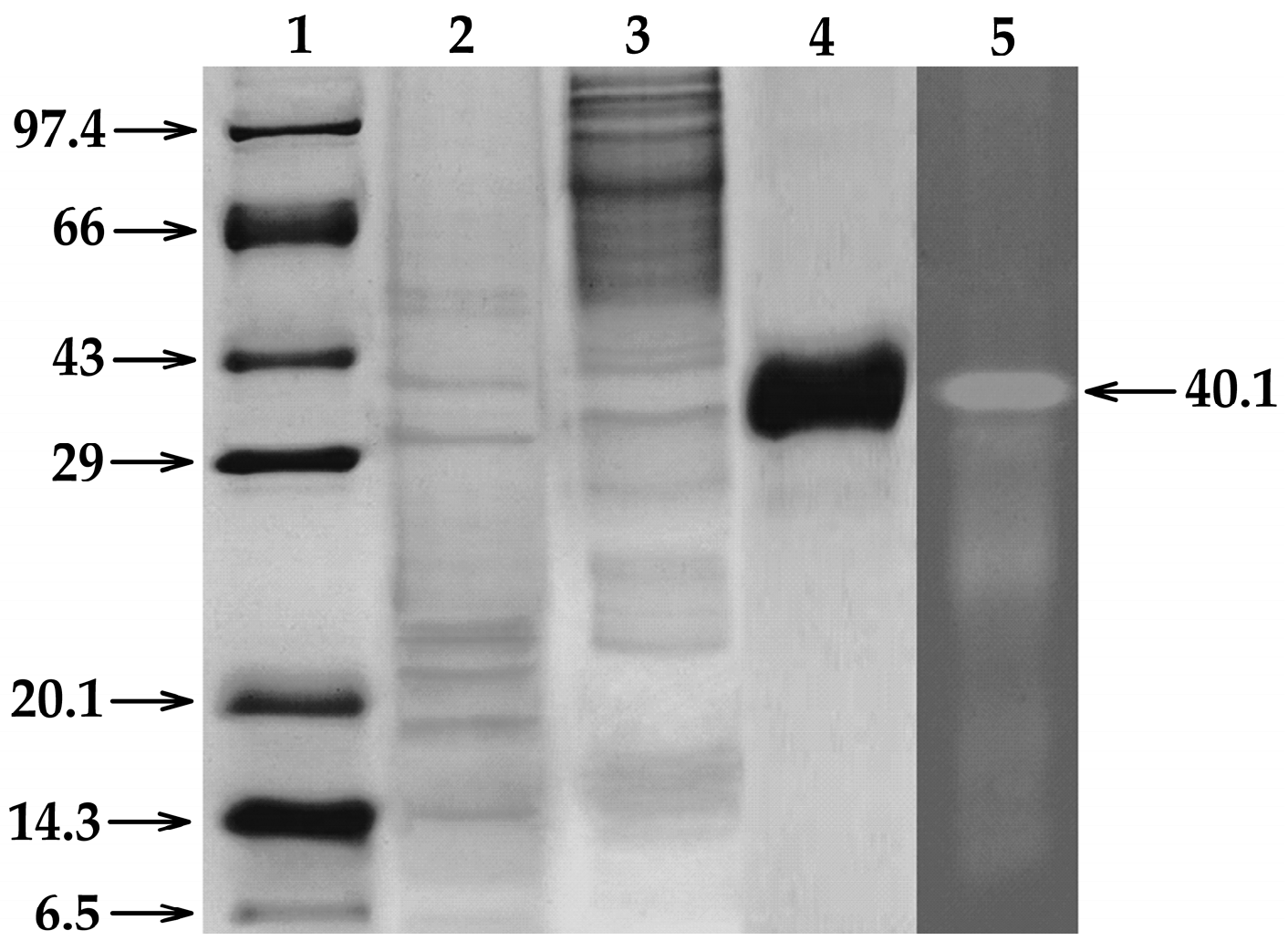
2.3. Expression and Purification of the Recombinant EglaRR01
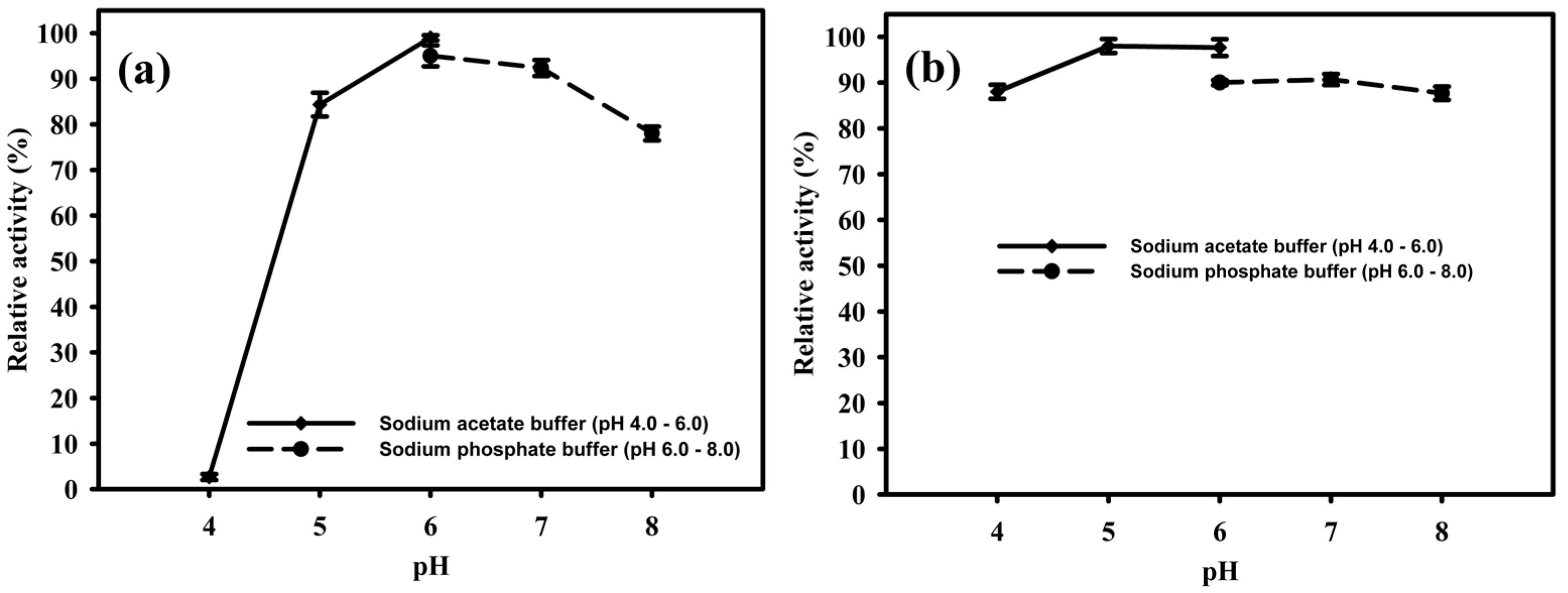
2.4. Temperature and pH Influence on the Activity of EglaRR01
2.5. Production of Bioethanol from Red Rice Compost
3. Discussion
4. Materials and Methods
4.1. Extraction and Purification of DNA from Environmental Samples
4.2. Construction of Metagenomic DNA Library and Screening
4.3. Sequence and Phylogenetic Analyses
4.4. Expression and Purification of a Recombinant Endoglucanase
4.5. Endoglucanase Activity Assay
4.6. CMC-Zymogram Analysis
4.7. Biochemical Characterization of Endoglucanase EglaRR01
4.8. Saccharification of Red Rice Residues by Endoglucanase EglaRR01
4.9. Bioconversion of Rice Straw Residues into Bioethanol Using Endoglucanase EglaRR01
4.9.1. Enzymatic Saccharification
4.9.2. Detoxification
4.9.3. Fermentation
4.9.4. Ethanol Estimation
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CMC | carboxymethylcellulose |
| pNPC | 4-nitrophenyl-β-d-cellobioside |
| CBM | carbohydrate binding motif |
| Ni-NTA | nickel-nitrilotriacetic acid |
| DNS | dinitrosalicylic acid |
| CFU | colony-forming unit |
References
- Lynd, L.R.; Weimer, P.J.; van Zyl, W.H.; Pretorius, I.S. Microbial cellulose utilization: Fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 2002, 66, 506–577. [Google Scholar] [CrossRef] [PubMed]
- Henrissat, B. A classification of glycosyl hydrolases based on amino-acid sequence similarities. Biochem. J. 1991, 280, 309–316. [Google Scholar] [PubMed]
- Collins, T.; Gerday, C.; Feller, G. Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol. Rev. 2005, 29, 3–23. [Google Scholar] [PubMed]
- Lynd, L.R.; van Zyl, W.H.; McBride, J.E.; Laser, M. Consolidated bioprocessing of cellulosic biomass: An update. Curr. Opin. Biotechnol. 2005, 16, 577–583. [Google Scholar] [PubMed]
- Steele, H.L.; Jaeger, K.E.; Daniel, R.; Streit, W.R. Advances in recovery of novel biocatalysts from metagenomes. J. Mol. Microbiol. Biotechnol. 2009, 16, 25–37. [Google Scholar] [PubMed]
- Rees, H.C.; Grant, S.; Jones, B.; Grant, W.D.; Heaphy, S. Detecting cellulase and esterase enzyme activities encoded by novel genes present in environmental DNA libraries. Extremophiles 2003, 7, 415–421. [Google Scholar] [PubMed]
- Grant, S.; Sorokin, D.Y.; Grant, W.D.; Jones, B.E.; Heaphy, S. A phylogenetic analysis of Wadi el Natrun soda lake cellulase enrichment cultures and identification of cellulase genes from these cultures. Extremophiles 2004, 8, 421–429. [Google Scholar] [PubMed]
- Voget, S.; Steele, H.L.; Streit, W.R. Characterization of a metagenome-derived halotolerant cellulase. J. Biotechnol. 2006, 126, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Warnecke, F.; Luginbühl, P.; Ivanova, N.; Ghassemian, M.; Richardson, T.H.; Stege, J.T.; Cayouette, M.; McHardy, A.C.; Djordjevic, G.; Aboushadi, N.; et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 2007, 405, 560–565. [Google Scholar] [CrossRef] [PubMed]
- Rappe, M.S.; Giovannoni, S.J. The uncultured microbial majority. Annu. Rev. Microbiol. 2003, 57, 369–394. [Google Scholar] [CrossRef] [PubMed]
- Daniel, R. The soil metagenome—A rich resource for the discovery of novel natural products. Curr. Opin. Biotechnol. 2004, 15, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Voget, S.; Leggewie, C.; Uesbeck, A.; Raasch, C.; Jaeger, K.E.; Streit, W.R. Prospecting for novel biocatalysts in a soil metagenome. Appl. Environ. Microbiol. 2003, 69, 6235–6242. [Google Scholar] [CrossRef] [PubMed]
- Teather, R.M.; Wood, P.J. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl. Environ. Microbiol. 1982, 43, 777–780. [Google Scholar] [PubMed]
- Zeng, R.; Xiong, P.; Wen, J. Characterization and gene cloning of a cold-active cellulase from a deep-sea psychrotrophic bacterium Pseudoalteromonas sp. DY3. Extremophiles 2006, 10, 79–82. [Google Scholar] [CrossRef] [PubMed]
- Schröder, C.; Blank, S.; Antranikian, G. First glycoside hydrolase family 2 enzymes from Thermus antranikianii and Thermus brockianus with β-glucosidase activity. Front. Bioeng. Biotechnol. 2015, 3, 1–10. [Google Scholar]
- Blank, S.; Schröder, C.; Schirrmacher, G.; Reisinger, C.; Antranikian, G. Biochemical characterization of a recombinant xylanase from Thermus brockianus, suitable for biofuel production. JSM Biotechnol. Biomed. Eng. 2014, 2, 1–10. [Google Scholar]
- Zhang, J.; Shi, H.; Xu, L.; Zhu, X.; Li, X. Site-directed Mutagenesis of a hyperthermophilic endoglucanase Cel12B from Thermotoga maritime based on rational design. PLoS ONE 2015, 10, e0133824. [Google Scholar]
- Lambertz, C.; Garvey, M.; Klinger, J.; Heesel, D.; Klose, H.; Fischer, R.; Commandeur, U. Challenges and advances in the heterologous expression of cellulolytic enzymes: A review. Biotechnol. Biofuels 2014, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Zhou, H.; Zhang, J.; Zhang, L.; Geng, A.; Liu, F.; Zhao, G.; Wang, S.; Zhou, Z.; Yan, X. Insight into dominant cellulolytic bacteria from two biogas digesters and their glycoside hydrolase genes. PLoS ONE 2015, 10, e0133824. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.H.P.; Lynd, L.R. Toward an aggregated understanding of enzymatic hydrolysis of cellulose: Noncomplexed cellulase systems. Biotechnol. Bioeng. 2004, 88, 797–824. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.T.; Thayer, D.W. The cellulase system of a Cytophaga species. Can. J. Microbiol. 1977, 23, 1285–1292. [Google Scholar] [CrossRef] [PubMed]
- Wood, T.M. The cellulase of Fusarium solani. Purification and specificity of the β-(1→4)-glucanase and the β-d-glucosidase components. Biochem. J. 1971, 121, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Baba, Y.; Sumitani, J.; Tani, S.; Kawaguchi, T. Characterization of Aspergillus aculeatus β-glucosidase 1 accelerating cellulose hydrolysis with Trichoderma cellulase system. Appl. Ind. Microbiol. Express 2015, 5, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Béra-Maillet, C.; Arthaud, L.; Abad, P.; Rosso, M.N. Biochemical characterization of MI-ENG1 a family 5 endoglucanase secreted by the root-knot nematode Meloidogyne incognita. Eur. J. Biochem. 2000, 267, 3255–3263. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Gruninger, R.J.; Qi, M.; Paterson, L.; Forster, R.J.; Teather, R.M.; McAllister, T.A. Cloning and identification of novel hydrolase genes from a dairy cow rumen metagenomic library and characterization of a cellulase gene. BMC Res. Notes 2012, 5, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Dong, D.; Wang, H.; Müller, K.; Qin, Y.; Wang, H.; Wu, W. Metagenomic analysis of microbial consortia enriched from compost: New insights into the role of Actinobacteria in lignocellulose decomposition. Biotechnol. Biofuels 2016, 9. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Antizar-Ladislao, B.; Khraisheh, M. Bioconversion of municipal solid waste to glucose for bio-ethanol production. Bioprocess Biosyst. Eng. 2007, 30, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.J.; Onkarapp, A.R.; Shobha, K.S. Comparative study of ethanol production from microbial pretreated agricultural residues. J. Appl. Sci. Environ. Manag. 2007, 11, 137–141. [Google Scholar]
- Kovacs, K.; Marcelli, S.; Szakacs, G.; Zacchi, G. Enzymatic hydrolysis of steam-preatreaated lignocellulosic materials with Trichoderma atroviride enzymes produced in-house. Biotechnol. Biofuels 2009, 2. [Google Scholar] [CrossRef] [PubMed]
- Rabelo, S.C.; Filho, R.M.; Costa, A.C. Lime pretreatment of sugarcane bagasse for bioethanol production. Appl. Biochem. Biotechnol. 2009, 53, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Yoswathana, N.; Phuriphipat, P. Bioethanol, production from rice straw. Energy Res. J. 2010, 1, 26–31. [Google Scholar] [CrossRef]
- Lynd, L.R.; Wyman, C.E.; Gerngross, T.U. Biocommodity engineering. Biotechnol. Prog. 1999, 15, 777–793. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Bruns, M.A.; Tiedje, J.M. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 1996, 62, 316–322. [Google Scholar] [PubMed]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Her, S.; Lee, H.S.; Choi, S.J.; Choi, S.W.; Choi, H.J.; Yoon, S.S.; Oh, D.H. Cloning and sequencing of β-1,4-endoglucanase gene (celA) from Pseudomonas sp. YD-15. Lett. Appl. Microbiol. 1999, 29, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Chavez, R.; Schachter, K.; Navarro, C.; Peirano, A.; Aguirre, C.; Bull, P.; Eyzaguirre, J. Differences in expression of two endoxylanase genes (xynA and xynB) from Penicillium purpurogenum. Gene 2002, 293, 161–168. [Google Scholar] [CrossRef]
- Yeh, Y.F.; Chang, S.C.; Kuo, H.W.; Tong, C.G.; Yu, S.M.; Ho, T.H. A metagenomic approach for the identification and cloning of an endoglucanase from rice straw compost. Gene 2013, 519, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, H.; Bajaj, K.L.; Arneja, J.S. Biochemical studies on bioconversion of rice straw to ethanol. Ind. J. Ecol. 1998, 25, 62–65. [Google Scholar]
- Caputi, A.J.; Ueda, M.; Brown, T. Spectrophotometric determination of ethanol in wine. Am. J. Enol. Vitic. 1968, 19, 160–165. [Google Scholar]
- Sample Availability: Samples of the purified EglaRR01 cellulase are available from the authors.

| Substrate | Specific Activity (U/mg) |
|---|---|
| β-d-glucan (Barley) | 1487.9 |
| CMC | 1002.5 |
| Avicel | 0 |
| Cellulose fiber | 5.8 |
| Cellobiose | 0 |
| Xylan-birchwood | 500.4 |
| Lichenan | 433.9 |
| Laminarin | 6.5 |
| Filter paper | 2.9 |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meneses, C.; Silva, B.; Medeiros, B.; Serrato, R.; Johnston-Monje, D. A Metagenomic Advance for the Cloning and Characterization of a Cellulase from Red Rice Crop Residues. Molecules 2016, 21, 831. https://doi.org/10.3390/molecules21070831
Meneses C, Silva B, Medeiros B, Serrato R, Johnston-Monje D. A Metagenomic Advance for the Cloning and Characterization of a Cellulase from Red Rice Crop Residues. Molecules. 2016; 21(7):831. https://doi.org/10.3390/molecules21070831
Chicago/Turabian StyleMeneses, Carlos, Bruna Silva, Betsy Medeiros, Rodrigo Serrato, and David Johnston-Monje. 2016. "A Metagenomic Advance for the Cloning and Characterization of a Cellulase from Red Rice Crop Residues" Molecules 21, no. 7: 831. https://doi.org/10.3390/molecules21070831








