Interaction of Di-2-pyridylketone 2-pyridine Carboxylic Acid Hydrazone and Its Copper Complex with BSA: Effect on Antitumor Activity as Revealed by Spectroscopic Studies
Abstract
:1. Introduction
2. Results and Discussion
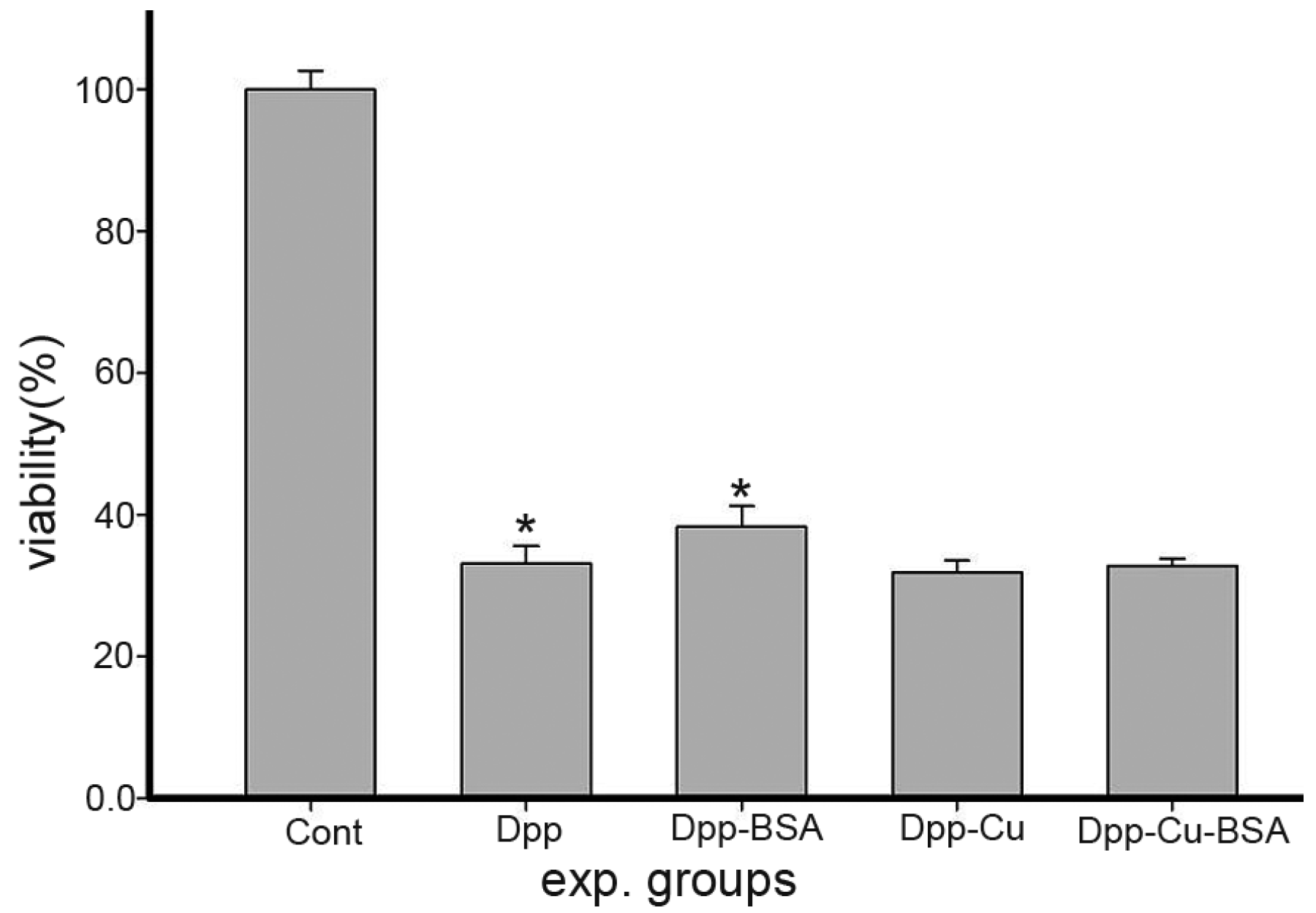
2.1. BSA Mediated Cytotoxicity of DPPCAH and Its Copper Complex (DPPCAH-Cu)
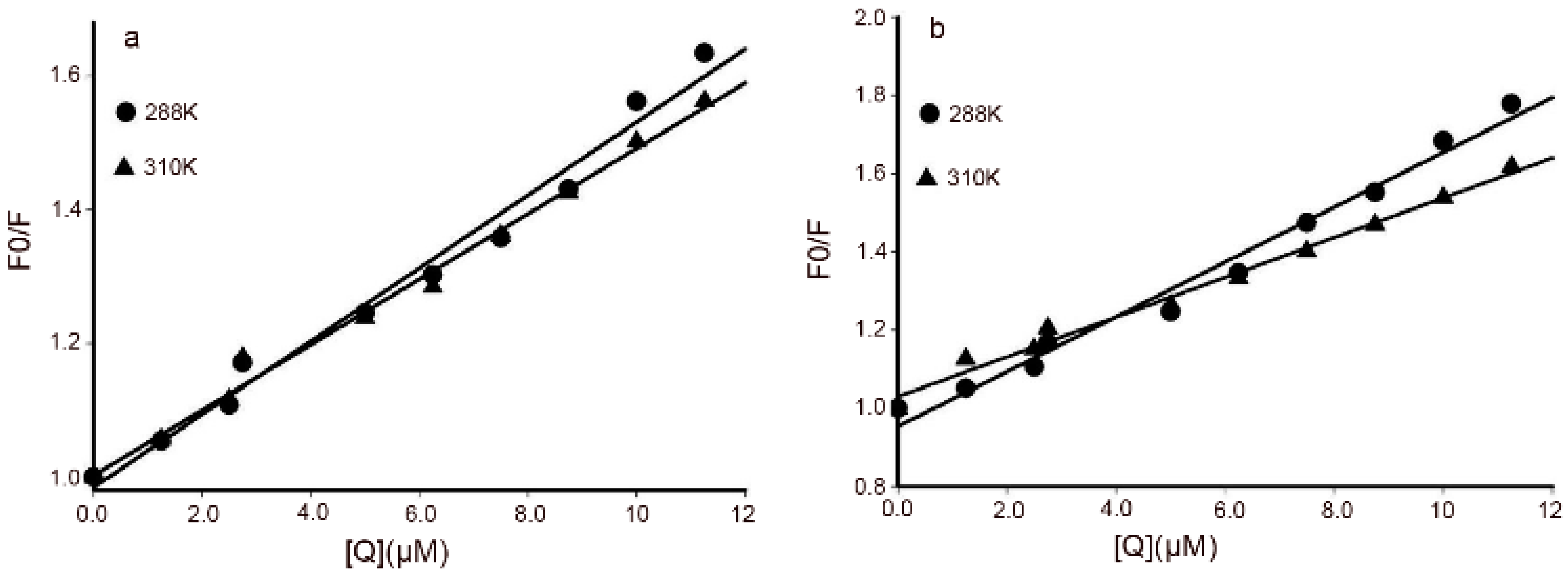
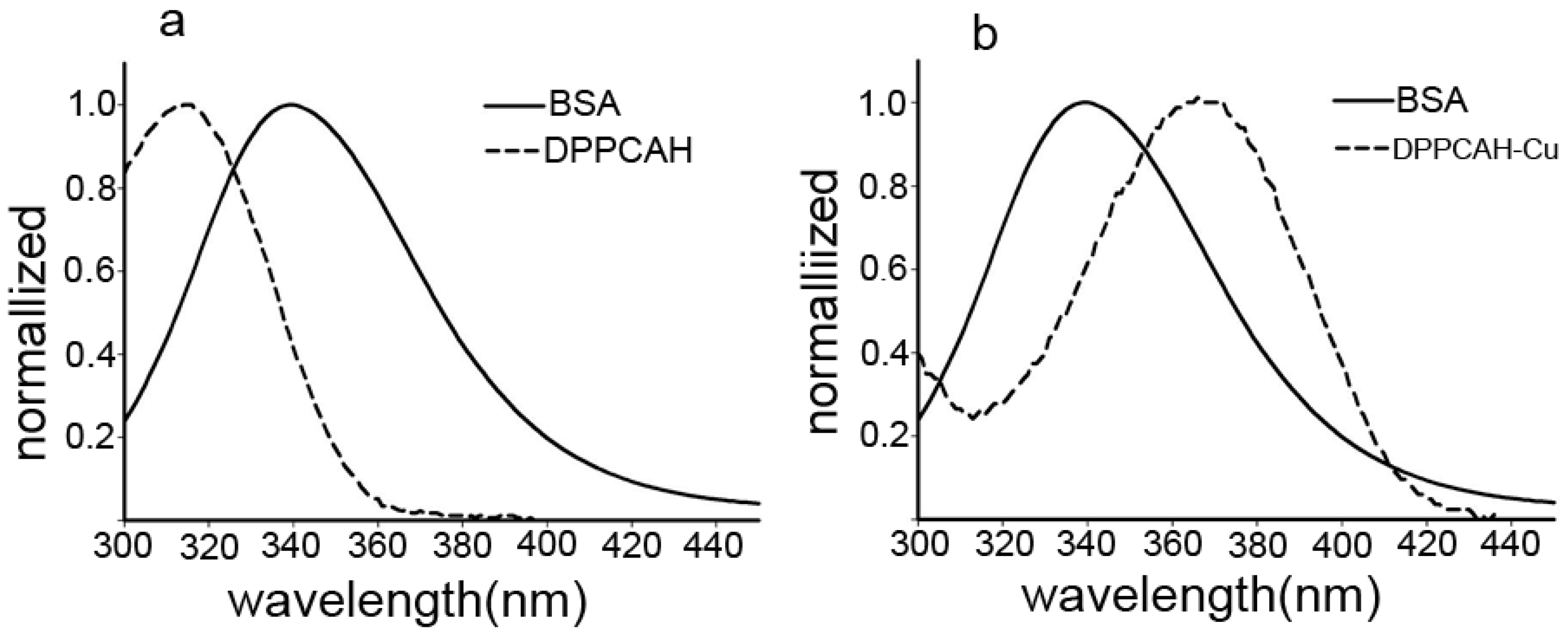
2.2. Interactions of DPPCAH and DPPCAH-Cu with BSA
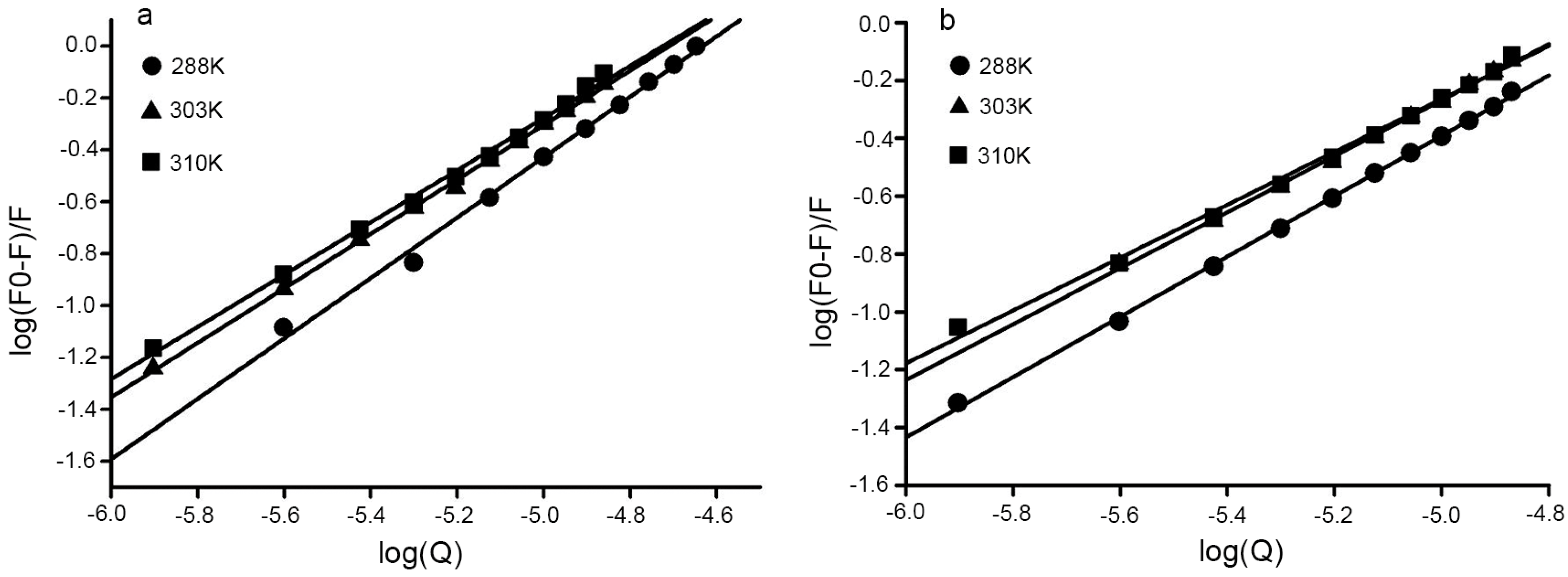
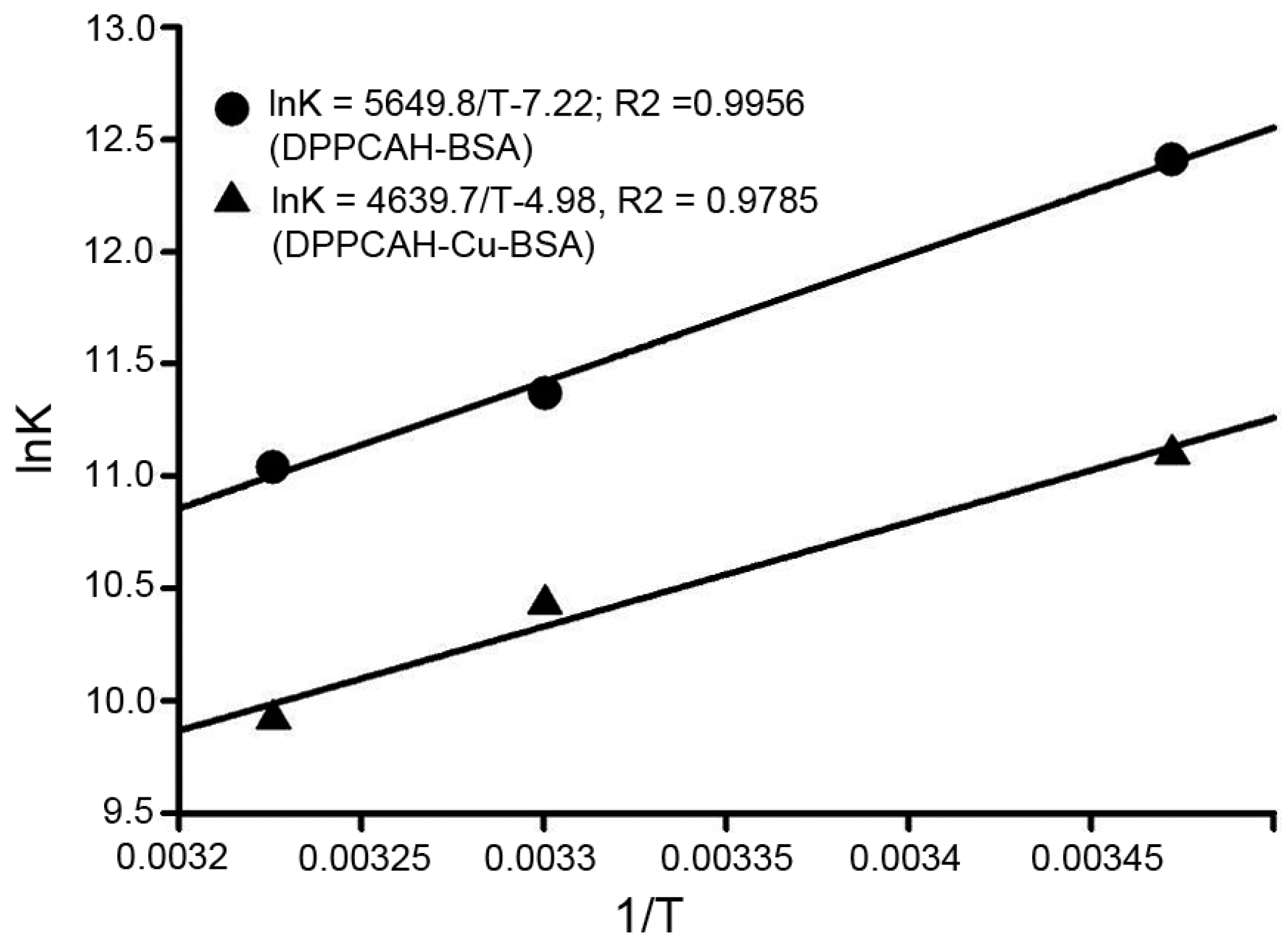
2.3. Binding Sites and Binding Constants
2.4. Binding Sites and Binding Constants
2.5. Binding Forces
2.6. Energy Transfer between DPPCAH (DPPCAH-Cu) and BSA
2.7. Conformational Investigation by Synchronous Fluorescence
2.8. Circular Dichroism Spectra
2.9. Molecular Docking
3. Materials and Methods
3.1. General Information
3.2. Cytotoxicity Assay (MTT Assay)
3.3. UV–Vis Spectrophotometric Measurements
3.4. Fluorescence Measurements
3.5. Measurements of CD Spectra
3.6. Molecular Docking
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| BSA | Bovine serum albumin |
| DPPCAH | Di-2-pyridylketone 2-pyridinecarboxylic acid hydrazone |
| DPPCAH-Cu | Copper complex of di-2-pyridylketone 2-pyridinecarboxylic acid hydrazone |
| FRET | Fluorescence resonance energy transfer |
References
- Chen, Z.Q.; Zhang, D.H.; Yue, F.; Zheng, M.H.; Kovacevic, Z.; Richardson, D.R. The Iron Chelators Dp44mT and DFO Inhibit TGF-β-induced Epithelial-Mesenchymal Transition via Up-Regulation of N-Myc Downstream-regulated Gene 1 (NDRG1). J. Biol. Chem. 2012, 287, 17016–17028. [Google Scholar] [CrossRef] [PubMed]
- Gupte, A.; Mumper, R.J. Elevated copper and oxidative stress in cancer cells as a target for cancer treatment. Cancer Treat. Rev. 2009, 35, 32–46. [Google Scholar] [CrossRef] [PubMed]
- Pahl, P.M.; Horwitz, L.D. Cell permeable iron chelators as potential cancer chemotherapeutic Agents. Cancer Investig. 2005, 23, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Knox, J.J.; Hotte, S.J.; Kollmannsberger, C.; Winquist, E.; Fisher, B.; Eisenhauer, E.A. Phase II study of Triapine in patients with metastatic renal cell carcinoma: A trial of the National Cancer Institute of Canada Clinical Trials Group (NCICIND.161). Investig. New Drugs 2007, 25, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.F.; Li, C.P.; Sun, X.Z.; Zhu, Z.F.; Liu, Y.X.; Yuan, Y.B.; Li, S.S.; Li, C.Z. The antitumor mechanism of di-2-pyridylketone 2-pyridinecarboxylic acid hydrazone and its copper complex in ROS generation and topoisomerase inhibition, and hydrazone involvement in oxygen-catalytic iron mobilization. Int. J. Oncol. 2015, 47, 1854–1862. [Google Scholar] [PubMed]
- Filipovic, N.; Borrmann, H.; Todorovic, T.; Borna, M.; Spasojević, V.; Sladić, D.; Novaković, I.; Andjelković, K. Copper(II) complexes of N-heteroaromatic hydrazones: Synthesis, X-ray structure, magnetic behavior, and antibacterial activity. Inorg. Chim. Acta 2009, 362, 1996–2000. [Google Scholar] [CrossRef]
- Fatfat, M.; Merhi, R.A.; Rahal, O.; Stoyanovsky, D.A.; Zaki, A.; Haidar, H.; Kagan, V.E.; Gali-Muhtasib, H.; Machaca, K. Copper chelation selectively kills colon cancer cells through redox cycling and generation of reactive oxygen species. BMC Cancer 2014, 14, 527. [Google Scholar] [CrossRef] [PubMed]
- Cuin, A.; Massabni, A.C.; Pereira, G.A.; Leite, C.Q.; Pavan, F.R.; Sesti-Costa, R.; Heinrich, T.A.; Costa-Neto, C.M. 6-Mercaptopurine complexes with silver and gold ions: Anti-tuberculosis and anticancer activities. Biomed. Pharmacother. 2011, 65, 334–338. [Google Scholar] [CrossRef] [PubMed]
- Navarro, M.; Castro, W.; González, S.; Abad, M.J.; Taylor, P. Synthesis and anticancer activity of gold(I)-chloroquine complexes. J. Mex. Chem. Soc. 2013, 57, 220–229. [Google Scholar]
- Li, H.H.; Bu, X.Y.; Lu, J.; Xu, C.Z.; Wang, X.L.; Yang, X.D. Interaction study of ciprofloxacin with human telomeric DNA by spectroscopy molecular docking. Spectrochim. Acta A 2013, 107, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Flarakos, J.; Morand, K.L.; Vouros, P. High-Throughput Solution-Based Medicinal Library Screening against Human Serum Albumin. Anal. Chem. 2005, 77, 1345–1353. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.; Zhang, L.; Sun, Y.; Diao, J.X.; Yang, X.L.; Sun, Y.; Zhang, L. Specificity and affinity of phenosafranine protein adduct: Insights from biophysical aspects. J. Lumin. 2012, 132, 629–635. [Google Scholar] [CrossRef]
- Merlot, A.M.; Sahni, S.; Lane, D.J.R.; Fordham, A.M.; Pantarat, N.; Hibbs, D.E.; Richardson, V.; Doddareddy, M.R.; Jennifer, A.; Huang, M.L.H.; et al. Potentiating the cellular targeting and anti-tumor activity of Dp44mT via binding to human serum albumin: Two saturable mechanisms of Dp44mT uptake by cells. Oncotarget 2015, 6, 10374–10398. [Google Scholar] [CrossRef] [PubMed]
- Merlot, A.M.; Kalinowski, D.S.; Richardson, D.R. Unraveling the mysteries of serum albumin-more than just a serum protein. Front. Physiol. 2014, 5, 299. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, Y.; Maeda, H. A new concept for macromolecular therapeutics in cancer chemotherapy: Mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Res. 1986, 46, 6387–6392. [Google Scholar] [PubMed]
- Carmeliet, P.; Jain, R.K. Angiogenesis in cancer and other diseases. Nature 2000, 407, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.L.; Yao, N.N.; Xu, H.R.; Wang, T.S.; Li, G.Y.; Li, Z.Q. Characterization of the interaction between eupatorin and bovine serum albumin by spectroscopic and molecular modeling methods. Int. J. Mol. Sci. 2013, 14, 14185–14203. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.X.; Li, X.; Snyder, M. Investigating metabolite-protein interactions: An overview of available techniques. Methods 2012, 57, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Klajnert, B.; Stanisławska, L.; Bryszewsk, M.; PaClecz, B. Interactions between PAMAM dendrimers and bovine serum albumin. Biochim. Biophys. Acta 2003, 1648, 115–126. [Google Scholar] [CrossRef]
- Li, Q.; Yang, W.Y.; Qu, L.L.; Qi, H.Y.; Huang, Y.; Zhang, Z. Interaction of Warfarin with Human Serum Albumin and Effect of Ferulic Acid on the Binding. J. Spectrosc. 2014, 2014, 834501. [Google Scholar] [CrossRef]
- Ross, P.D.; Subramanian, S. Thermodynamics of protein association reactions: Forces contributing to stability. Biochemistry 1981, 20, 3096–3102. [Google Scholar] [CrossRef] [PubMed]
- Du, H.Y.; Xiang, J.F.; Zhang, Y.Z.; Tang, Y.L. A spectroscopic and molecular modeling study of sinomenine binding to transferring. Bioorg. Med. Chem. Lett. 2007, 17, 1701–1704. [Google Scholar] [CrossRef] [PubMed]
- Li, D.J.; Zhu, J.F.; Jin, J.; Yao, X.J. Studies on the binding of nevadensin to human serum albumin by molecular spectroscopy and modeling. J. Mol. Struct. 2007, 846, 34–41. [Google Scholar] [CrossRef]
- Timaseff, S.N. Thermodynamics of Protein Interactions; Pergamon Press: Oxford, UK, 1972; pp. 511–519. [Google Scholar]
- Mohammadi, F.; Moeeni, M. Study on the interactions of trans-resveratrol and curcumin with bovine α-lactalbumin by spectroscopic analysis and molecular docking. Mater. Sci. Eng. C. 2015, 50, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.; Khan, A.Y.; Kumar, G.S. Interaction of the anticancer plant alkaloid sanguinarine with bovine serum albumin. PLoS ONE 2011, 6, e18333. [Google Scholar] [CrossRef] [PubMed]
- Varlan, A.; Hillebrand, M. Bovine and human serum albumin interactions with 3-carboxyphenoxathiin studied by fluorescence and circular dichroism spectroscopy. Molecules 2010, 15, 3905–3919. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.Z.; Lin, Z.; Yang, L.J.; Wang, C.; Bai, C.L. Investigation of the interaction between acridine orange and bovine serum albumin. Talanta 1998, 47, 1223–1229. [Google Scholar] [CrossRef]
- Tan, M.X.; Liang, W.J.; Luo, X.J.; Gu, Y.Q. Fluorescence Spectroscopy Study on the Interaction between Evodiamine and Bovine Serum Albumin. J. Chem. 2013, 2013, 308054. [Google Scholar] [CrossRef]
- Sułkowska, A.; Maciaźeka, M.; Rŏwnicka, J.; Bojko, B.; Pentak, D.; Sułkowski, W.W. Effect of temperature on the methotrexate-BSA interaction: Spectroscopic study. J. Mol. Struct. 2007, 834, 162–169. [Google Scholar] [CrossRef]
- Ibrahim, N.; Ibrahim, H.; Kim, S.; Nallet, J.P.; Nepveu, F. Interactions between antimalarial indolone-N-oxide derivatives and human serum albumin. Biomacromolecules 2010, 11, 3341–3351. [Google Scholar] [CrossRef] [PubMed]
- Sułkowska, A. Interaction of drugs with bovine and human serumalbumin. J. Mol. Struct. 2002, 614, 227–232. [Google Scholar] [CrossRef]
- Vekshin, N.L. Separation of the tyrosine and tryptophan components of fluorescence using synchronous scanning method. Biofizika 1996, 41, 1176–1179. [Google Scholar] [PubMed]
- Ghisaidoobe, A.B.T.; Chung, S.J. Intrinsic tryptophan fluorescence in the detection and analysis of proteins: A focus on Förster Resonance Energy Transfer techniques. Int. J. Mol. Sci. 2014, 15, 22518–22538. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.G.; Liu, R.; Jiang, X.H. Spectroscopic studies on the interaction between tetrandrine and two serum albumins by chemometrics methods. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 115, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Chi, Z.X.; Liu, R.T. Phenotypic Characterization of the Binding of Tetracycline to Human Serum Albumin. Biomacromolecules 2011, 12, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Kandagal, P.B.; Ashoka, S.; Seetharamappa, J.; Shaikh, S.M.T.; Jadegoud, Y.; Ijare, O.B. Study of the interaction of an anticancer drug with human and bovine serum albumin: Spectroscopic approach. J. Pharm. Biomed. Anal. 2006, 41, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Liao, Z.X.; Yu, X.Y.; Yao, Q.; Yi, P.G. Interaction between pirenoxine and bovine serum albumin in aqueous Solution. Spectrochim. Acta Part A. 2014, 129, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Perez-Iratxeta, C.; Andrade-Navarro, M.A. K2D2: Estimation of protein secondary structure from circular dichroism spectra. BMC Struct. Biol. 2008, 8, 25. [Google Scholar] [CrossRef] [PubMed]
- Bujacz, A.; Zielinski, K.; Sekula, B. Structural studies of bovine, equine, and leporine serum albumin complexes with naproxen. Proteins 2014, 82, 2199–2208. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Petitpas, I.; Petersen, C.E.; Ha, C.E.; Bhattacharya, A.A.; Zunszain, P.A.; Ghuman, J.; Bhagavan, N.V.; Curry, S. Structural basis of albumin-thyroxine interactions and familial dysalbuminemic hyperthyroxinemia. Proc. Natl. Acad. Sci. USA 2003, 100, 6440–6445. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System; DeLano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds are available from the authors.




| System | Δ𝐻 (kJ·mol−1) | ΔS J/mol·K | ΔG (kJ·mol−1) |
|---|---|---|---|
| BSA-DPPCAH | −46.97 | −60.0 | −29.69 |
| BSA-DPPCAH-Cu | −38.57 | −41.5 | −26.61 |
| System | J (cm3·L·mol−1) | E (%) | R0 (nm) | r (nm) |
|---|---|---|---|---|
| BSA-DPPCAH | 7.424 × 1013 | 0.297 | 2.35 | 2.04 |
| BSA-DPPCAH-Cu | 1.699 × 1014 | 0.288 | 2.70 | 2.32 |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Huang, T.; Fu, Y.; Liu, Y.; Zhou, S.; Qi, Z.; Li, C. Interaction of Di-2-pyridylketone 2-pyridine Carboxylic Acid Hydrazone and Its Copper Complex with BSA: Effect on Antitumor Activity as Revealed by Spectroscopic Studies. Molecules 2016, 21, 563. https://doi.org/10.3390/molecules21050563
Li C, Huang T, Fu Y, Liu Y, Zhou S, Qi Z, Li C. Interaction of Di-2-pyridylketone 2-pyridine Carboxylic Acid Hydrazone and Its Copper Complex with BSA: Effect on Antitumor Activity as Revealed by Spectroscopic Studies. Molecules. 2016; 21(5):563. https://doi.org/10.3390/molecules21050563
Chicago/Turabian StyleLi, Cuiping, Tengfei Huang, Yun Fu, Youxun Liu, Sufeng Zhou, Zhangyang Qi, and Changzheng Li. 2016. "Interaction of Di-2-pyridylketone 2-pyridine Carboxylic Acid Hydrazone and Its Copper Complex with BSA: Effect on Antitumor Activity as Revealed by Spectroscopic Studies" Molecules 21, no. 5: 563. https://doi.org/10.3390/molecules21050563





