Eremophilane Sesquiterpenes from a Deep Marine-Derived Fungus, Aspergillus sp. SCSIOW2, Cultivated in the Presence of Epigenetic Modifying Agents
Abstract
:1. Introduction
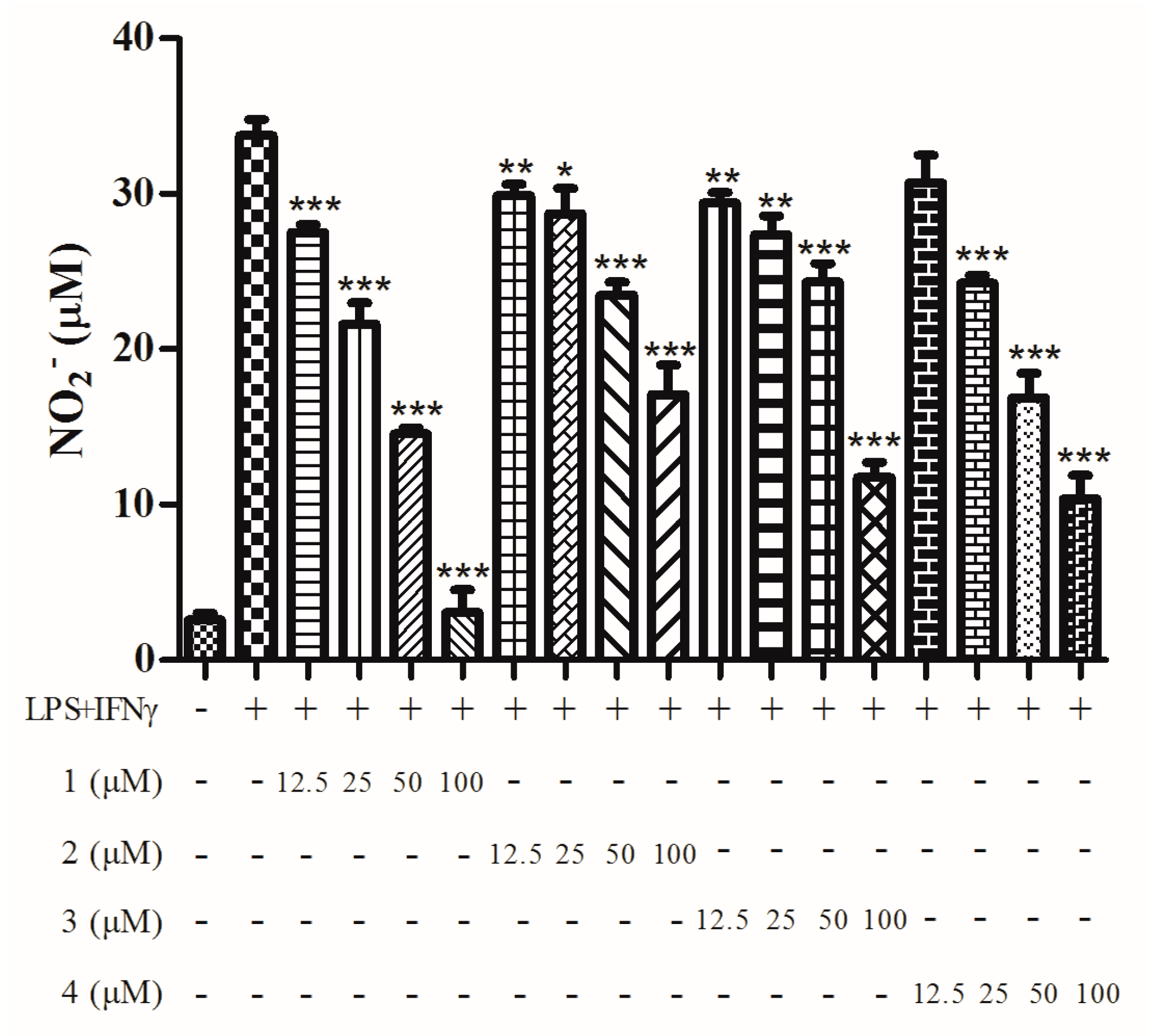
2. Results
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Strain
4.3. Fermentation, Extraction, and Isolation
4.4. Quantum Chemical ECD Calculations
4.5. Cell Culture Condition
4.6. Nitric Oxide Inhibitory Assay
4.7. MTT Assay
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 2012, 75, 311–335. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2012, 29, 144–222. [Google Scholar] [CrossRef] [PubMed]
- Scherlach, K.; Hertweck, C. Triggering cryptic natural product biosynthesis in microorganisms. Org. Biomol. Chem. 2009, 7, 1753–1760. [Google Scholar] [CrossRef] [PubMed]
- Shwab, E.K.; Bok, J.W.; Tribus, M.; Galehr, J.; Graessle, S.; Keller, N.P. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot. Cell 2007, 6, 1656–1664. [Google Scholar] [CrossRef] [PubMed]
- Cichewicz, R.H. Epigenome manipulation as a pathway to new natural product scaffolds and their congeners. Nat. Prod. Rep. 2010, 27, 11–22. [Google Scholar] [CrossRef] [PubMed]
- VanderMolen, K.M.; Darveaux, B.A.; Chen, W.L.; Swanson, S.M.; Pearce, C.J.; Oberlies, N.H. Epigenetic manipulation of a filamentous fungus by the proteasome-inhibitor bortezomib induces the production of an additional secondary metabolite. RSC Adv. 2014, 4, 18329–18335. [Google Scholar] [CrossRef] [PubMed]
- Wiliams, R.B.; Henrikson, J.C.; Hoover, A.R.; Lee, A.E.; Cichewicz, R.H. Epigenetic remodeling of the fungal secondary metabolome. Org. Biomol. Chem. 2008, 7, 1895–1897. [Google Scholar] [CrossRef] [PubMed]
- Vervoort, H.C.; Draskovic, M.; Crews, P. A novel dipeptide from a hawaiian marine sediment-derived fungus, Aspergillus insulicola. Org. Lett. 2011, 13, 410–413. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Shao, C.L.; Chen, M.; Liu, Q.A.; Wang, C.Y. Brominated resorcylic acid lactones from the marine-derived fungus Cochliobolus lunatus induced by histone deacetylase inhibitors. Tetrahedron Lett. 2014, 55, 4888–4891. [Google Scholar] [CrossRef]
- Wang, X.R.; Sena Filho, J.G.; Hoover, A.R.; King, J.B.; Ellis, T.K.; Powell, D.R.; Cichewicz, R.H. Chemical epigenetics alters the secondary metabolite composition of guttate excreted by an atlantic-forest-soil-derived Penicillium citreonigrum. J. Nat. Prod. 2010, 73, 942–948. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.M.; Wei, C.K.; Chuang, D.W.; El-Shazly, M.; Hsieh, C.T.; Asai, T.; Oshima, Y.; Hsieh, T.J.; Hwang, T.L.; Wu, Y.C.; et al. An epigenetic modifier enhances the production of anti-diabetic and anti-inflammatory sesquiterpenoids from Aspergillus sydowii. Bioorg. Med. Chem. 2013, 21, 3866–3872. [Google Scholar] [CrossRef] [PubMed]
- Asai, T.; Chung, Y.M.; Sakurai, H.; Ozeki, T.; Chang, F.R.; Yamashita, K.; Oshima, Y. Aromatic polyketide production in Cordyceps indigotica, an entomopathogenic fungus, induced by exposure to a histone deacetylase inhibitor. Org. Lett. 2012, 54, 2006–2009. [Google Scholar] [CrossRef] [PubMed]
- Asai, T.; Chung, Y.M.; Sakurai, H.; Ozeki, T.; Chang, F.R.; Wu, Y.C.; Yamashita, K.; Oshima, Y. Highly oxidized ergosterols and isariotin analogs from an entomopathogenic fungus, Gibellula formosana, cultivated in the presence of epigenetic modifying agents. Tetrahedron 2012, 68, 5817–5823. [Google Scholar] [CrossRef]
- Zhou, X.; Fang, P.Y.; Tang, J.Q.; Wu, Z.Q.; Li, X.F.; Li, S.M.; Wang, Y.; Liu, G.; He, Z.D.; Gou, D.M.; et al. A novel cyclic dipeptide from deep marine-derived fungus Aspergillus sp. SCSIOW2. Nat. Prod. Res. 2015, 24, 51–57. [Google Scholar]
- Sugawara, F.; Strobel, G.; Fisher, L.; Van Duyne, G.D.; Clardy, J. Bipolaroxin, a selective phytotoxin produced by Bipolaris cynodontis. Proc. Natl. Acad. Sci. USA 1985, 82, 8291–8294. [Google Scholar] [CrossRef] [PubMed]
- Hou, C.J.; Kulka, M.; Zhang, J.P.; Li, Y.M.; Guo, F.J. Occurrence and biological activities of eremophilane-type Sesquiterpenes. Mini-Rev. Midecinal Chem. 2014, 14, 664–677. [Google Scholar] [CrossRef]
- Aktan, F.; Henness, S.; Roufogalis, B.D.; Ammit, A.J. Gypenosides derived from Gynostemma pentaphyllum suppress NO synthesis in murine macrophages by inhibiting iNOS enzymatic activity and attenuating NF-κB-mediated iNOS protein expression. Nitric Oxide 2003, 8, 235–242. [Google Scholar] [CrossRef]
- Bogdan, C. Nitric oxide and the immune response. Nat. Immunol. 2001, 2, 907–916. [Google Scholar] [CrossRef] [PubMed]
- Lorsbach, R.B.; Murphy, W.J.; Lowenstein, C.J.; Snyder, S.H.; Russell, W. Expression of the nitric oxide synthase gene in mouse macrophages activated for tumor cell killing. Molecular basis for the synergy between interferon-gamma and lipopolysaccharide. J. Biol. Chem. 1993, 268, 1908–1913. [Google Scholar] [PubMed]
- Tötemeyer, S.; Sheppard, M.; Lloyd, A.; Roper, D.; Dowson, C.; Underhill, D.; Murray, P.; Maskell, D.; Bryant, C. IFN-γ enhances production of nitric oxide from macrophages via a mechanism that depends on nucleotide oligomerization domain-2. J. Immunol. 2006, 176, 4804–4810. [Google Scholar] [CrossRef] [PubMed]
- Van Meerloo, J.; Kaspers, G.J.; Cloos, J. Cell sensitivity assays: The MTT assay. Methods Mol. Biol. 2011, 731, 237–245. [Google Scholar] [PubMed]
- Hwang, B.Y.; Lee, J.H.; Koo, T.H.; Kim, H.S.; Hong, Y.S.; Ro, J.S.; Lee, K.S.; Lee, J.J. Furanoligularenone, an eremophilane from Ligularia fischeri, inhibits the LPS-induced production of nitric oxide and prostaglandin E2 in macrophage RAW264.7 cells. Planta Med. 2002, 68, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.H.; Shen, T.; Yang, X.; Zhao, H.; Li, X.; Xie, W.D. Sesquiterpenoids from Farfugium japonicum and their inhibitory activity on NO production in RAW264.7 Cells. Arch. Pharmacal Res. 2012, 35, 1153–1158. [Google Scholar] [CrossRef] [PubMed]
- Nugroho, E.A.; Morita, H. Circular dichroism calculation for natural products. J. Nat. Med. 2014, 68, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Cresset. Available online: http://www.cresset-group.com (accessed on 9 April 2016).
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision D.01; Gaussian Inc.: Wallingford, CT, USA, 2013. [Google Scholar]
- Bruhn, T.; Schaumlöffel, A.; Hemberger, Y.; Bringmann, G. SpecDis, Version 1.60; University of Wuerzburg: Wuerzburg, Germany, 2013. [Google Scholar]
- Bruhn, T.; Schaumlöffel, A.; Hemberger, Y.; Bringmann, G. SpecDis: Quantifying the comparison of calculated and experimental electronic circular dichroism spectra. Chirality 2013, 25, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Wang, L.; Liu, K. In vitro anti-inflammatory effects of arctigenin, a lignan from Arctium lappa L., through inhibiton on iNOS pathway. J. Ethnopharmacol. 2009, 122, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, L.Y.; Wu, Z.Q.; Yao, L.J.; Wu, Y.H.; Huang, L.; Liu, K.; Zhou, X.; Gou, D.M. Anthocyanin-rich fractions from red raspberries attenuate inflammation in both RAW264.7 macrophages and a mouse model of colitis. Sci. Rep. 2014. [Google Scholar] [CrossRef]
- Denizot, F.; Lang, R. Rapid colorimetric assay for cell growth and survival. Modifications to the tetrazolium dye proceduregiving improved sensitivity and reliability. J. Immunol. Methods 1986, 89, 271–277. [Google Scholar] [CrossRef]
- Sample Availability: Samples of the compounds 1–4 are available from the authors.
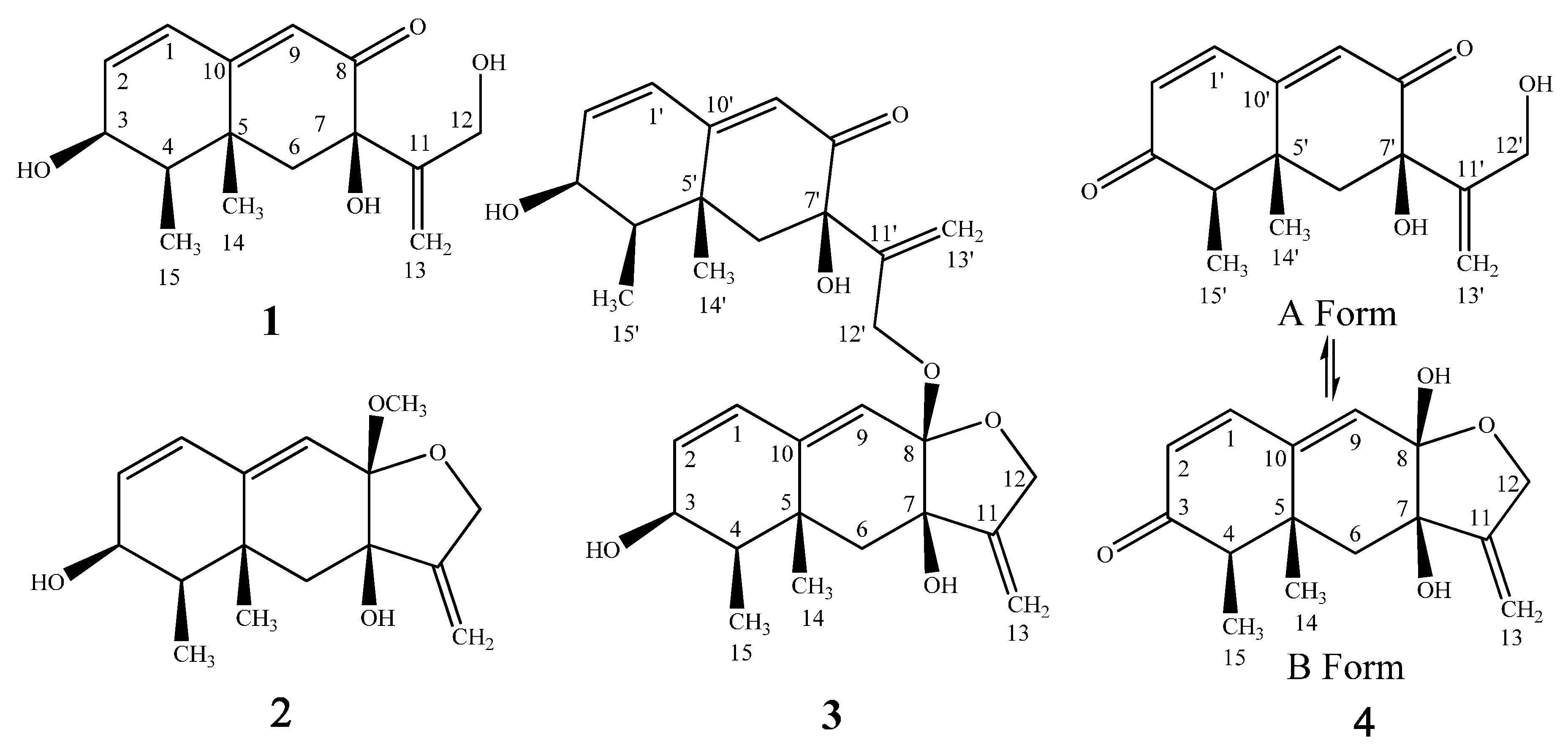
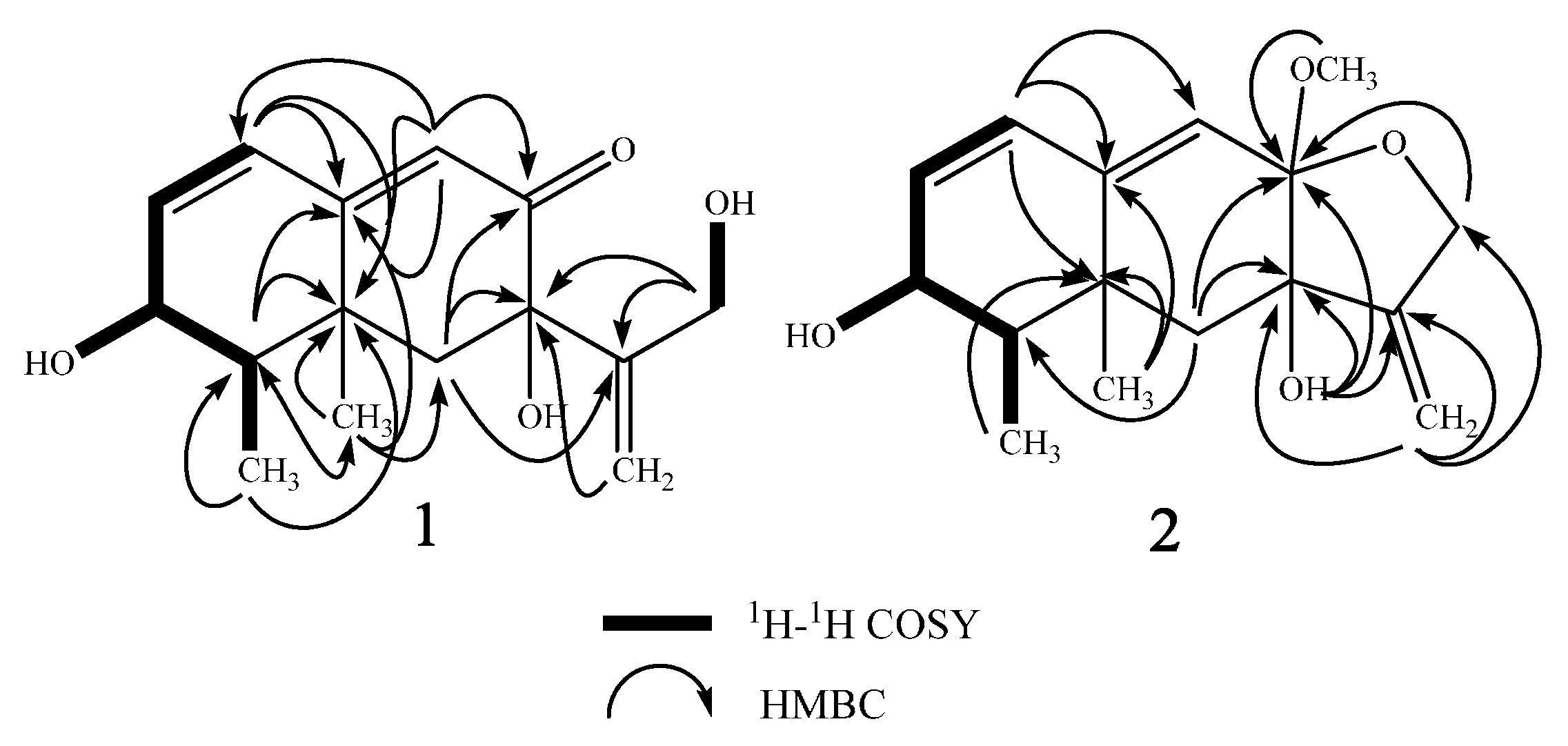




| Position | δH (Mult, J in Hz) b | δC c | 1H-1H COSY | HMBC | NOESY |
|---|---|---|---|---|---|
| 1 | 6.26 m d | 127.4 | 2 | C-2,3,5,9,10 | 2,9 |
| 2 | 6.24 m d | 139.0 | 1,3 | C-3,4,10 | 1 |
| 3 | 4.03 m | 66.0 | 2,4,3-OH | C-1,2 | 4, 15 |
| 4 | 1.57 m | 41.8 | 3,15 | C-5,10,14,15 | 3,6α |
| 5 | 35.9 | ||||
| 6α | 1.86 d (14.4) | 46.1 | 6β | C-4,5,7,8,10,11,14 | 4,12 |
| 6β | 1.91 d (14.4) | 6α | C-4,5,7,8,10,11,14 | 14,15,13a | |
| 7 | 75.9 | ||||
| 8 | 196.9 | ||||
| 9 | 5.76 s | 122.9 | C-1,5,7,10 | 1 | |
| 10 | 163.0 | ||||
| 11 | 153.7 | ||||
| 12 | 3.82 d (4.2) (2H) | 60.9 | 12-OH | C-7,11,13 | 6α, 13b |
| 13a | 5.18 d (1.8) | 109.3 | 13b | C-7,11,12 | 6β |
| 13b | 5.21 d (1.8) | 13a | C-7,11,12 | 12, 12-OH | |
| 14 | 1.35 s | 22.6 | C-4,5,6,10 | 6β,7-OH,15 | |
| 15 | 0.99 d (6.6) | 10.7 | 4 | C-3,4,5 | 3,3-OH,6β,14 |
| 3-OH | 4.88 d (5.4) | 3 | C-2,3,4 | 15 | |
| 7-OH | 5.30 s | C-6,7 | 14 | ||
| 12-OH | 4.37 t (4.2) | 12 | C-11,12 | 13b |
| Position | δH (Mult, J in Hz) b | δC c | 1H-1H COSY | HMBC | NOESY |
|---|---|---|---|---|---|
| 1 | 6.05 d (9.6) | 128.0 | 2 | C-3,5,9,10 | 2,9 |
| 2 | 5.85 dd (9.6, 4.8) | 132.2 | 1,3 | C-1,3,4,10 | 1 |
| 3 | 3.89 m | 66.6 | 2,4,3-OH | C-1,2,4,5,15 | 4,15 |
| 4 | 1.36 m | 43.1 | 3,15 | C-3,5,10,14,15 | 3 |
| 5 | 35.9 | ||||
| 6α | 1.32 d (13.8) | 44.7 | 6β | C-5,7,8,10,11,14, | 4,12 |
| 6β | 1.68 d (13.8) | 6α | 14,15,13a | ||
| 7 | 78.6 | ||||
| 8 | 101.4 | ||||
| 9 | 5.72 s | 117.4 | C-1,5,7,10 | 1,8-OCH3 | |
| 10 | 144.9 | ||||
| 11 | 154.7 | ||||
| 12α | 4.37 d (13.8) | 66.7 | 12β | C-7,8,11,13 | 6α,13b |
| 12β | 4.24 d (13.8) | 12α | 13b | ||
| 13a | 5.12 s | 104.2 | 13b | C-7,11,12 | 6β |
| 13b | 4.92 s | 13a | 12α,12β | ||
| 14 | 1.25 s | 20.8 | C-4,5,6,10 | 6β,7-OH,8-OCH3,15 | |
| 15 | 0.94 d (6.6) | 10.6 | 4 | C-3,4,5 | 3,3-OH,6β,14 |
| 3-OH | 4.59 d (4.2) | 3 | C-2,3,4 | 15 | |
| 7-OH | 4.24 s | C-6,7,8,11 | 14 | ||
| 8-OCH3 | 3.27 s | 48.1 | C-8 | 9,7-OH,14 |
| No. | 3 | 4 | ||||
|---|---|---|---|---|---|---|
| δH (Mult, J in Hz) b | δC c | HMBC | δH (Mult, J in Hz) b | δC c | HMBC | |
| 1 | 5.99 d (9.6) | 127.8 | C-2,3,5,9,10 | 7.11 d (9.6) | 144.3 | C-2,3,9,10 |
| 2 | 5.84 dd (9.6,4.8) | 132.3 | C-1,3,4,10 | 5.93 d (9.6) | 127.2 | C-1,4,10 |
| 3 | 3.88 m | 66.5 | C-1,2,4,5 | 200.3 | ||
| 4 | 1.32 m | 43.0 | C-5,6,10,14,15 | 2.34 m | 52.2 | C-3,5,6,14,15 |
| 5 | 35.8 | 39.1 | ||||
| 6α | 1.30 d (13.8) | 44.3 | C-4,5,14 | 1.62 d (13.8) | 44.1 | C-4,5,7,8 |
| 6β | 1.69 d (13.8) | 1.76 d (13.8) | 10,11,14 | |||
| 7 | 78.7 | 76.2 | ||||
| 8 | 101.6 | 98.6 | ||||
| 9 | 5.61 s | 117.7 | C-1,5,7,8,10 | 6.01 s | 130.8 | C-1,5,7,8 |
| 10 | 144.8 | 140.8 | ||||
| 11 | 154.2 | 154.3 | ||||
| 12 | 4.37 d (13.8) | 67.0 | C-8,11,13 | 4.35 d (14.4) d | 66.2 | C-7,8,11,13 |
| 12 | 4.24 d (13.8) | 4.40 d (14.4) d | ||||
| 13a | 5.12 brs | 104.5 | C-7,11 | 5.18 s | 104.9 | C-7,11,12 |
| 13b | 4.89 brs d | 4.99 s | ||||
| 14 | 1.22 s | 20.8 | C-4,5,6 | 1.11 s | 19.95 | C-4,5,6,10 |
| 15 | 0.92 d (7.2) | 10.6 | C-3,4,5 | 0.86 d (6.6) | 7.09 | C-3,4,5 |
| 1′ | 6.26 m d | 139.0 | C-3′,5′,9′,10′ | 7.30 d (10.2) | 142.6 | C-3′,5′,9′,10′ |
| 2′ | 6.25 m d | 127.4 | C-1′,3′,4′, 10′ | 6.22 d (10.2) d | 131.5 | C-1′,4′,10′ |
| 3′ | 3.96 m | 66.1 | 200.0 | |||
| 4′ | 1.57 m | 41.8 | C-5′,6′,10′,14′,15′ | 2.70 m | 51.9 | C-3′,5′,6′,10′ |
| 5′ | 35.9 | 40.0 | 14′,15′ | |||
| 6′α | 1.91 s (2H) | 45.5 | C-7′,8′,10′ | 2.28 d (14.4) | 45.4 | C-5′,7′,8′ |
| 6′β | 11′,14′ | 1.92 d (14.4) | 10′,14′ | |||
| 7′ | 75.8 | 75.1 | ||||
| 8′ | 196.7 | 196.4 | ||||
| 9′ | 5.74 s | 122.9 | C-5′,7′,10′ | 6.22 s d | 127.5 | C-1′,5′,7′,10′ |
| 10′ | 163.2 | 158.9 | ||||
| 11′ | 149.6 | 152.7 | ||||
| 12′ | 4.08 s (2H) | 61.4 | C-8,6′,11′,13′ | 3.90 d (4.8) | 61.0 | C-7′,11′,13′ |
| 13′a | 5.28 brs (2H) | 112.5 | C-7′,11′,12′ | 5.26 s | 110.0 | C-7′,11′,12′ |
| 13′b | 5.23 s | |||||
| 14′ | 1.22 s | 22.6 | C-6′ | 1.23 s | 21.8 | C-4′,5′,6′,10′ |
| 15′ | 0.92 d (7.2) | 10.7 | C-3′,4′,5′ | 0.98 d (6.6) | 7.09 | C-3′,4′,5′ |
| 3-OH | 4.60 d (5.4) | C-2,3,4 | ||||
| 7-OH | 3.59 d (1.2) | C-6,7,8 | 6.54 s | C-7,8 | ||
| 8-OH | 4.36 s | C-7,8,9 | ||||
| 3′-OH | 4.88 d (6.6) d | C-3′,4′ | ||||
| 7′-OH | 5.39 s | C-11′ | 5.60 s | C-6′,7′,8′ | ||
| 12′-OH | 4.81 t (4.8) | C-12′ | ||||
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Li, M.; Tang, J.; Li, X. Eremophilane Sesquiterpenes from a Deep Marine-Derived Fungus, Aspergillus sp. SCSIOW2, Cultivated in the Presence of Epigenetic Modifying Agents. Molecules 2016, 21, 473. https://doi.org/10.3390/molecules21040473
Wang L, Li M, Tang J, Li X. Eremophilane Sesquiterpenes from a Deep Marine-Derived Fungus, Aspergillus sp. SCSIOW2, Cultivated in the Presence of Epigenetic Modifying Agents. Molecules. 2016; 21(4):473. https://doi.org/10.3390/molecules21040473
Chicago/Turabian StyleWang, Liyan, Mengjie Li, Jianqiang Tang, and Xiaofan Li. 2016. "Eremophilane Sesquiterpenes from a Deep Marine-Derived Fungus, Aspergillus sp. SCSIOW2, Cultivated in the Presence of Epigenetic Modifying Agents" Molecules 21, no. 4: 473. https://doi.org/10.3390/molecules21040473





