Yeast Extract and Silver Nitrate Induce the Expression of Phenylpropanoid Biosynthetic Genes and Induce the Accumulation of Rosmarinic Acid in Agastache rugosa Cell Culture
Abstract
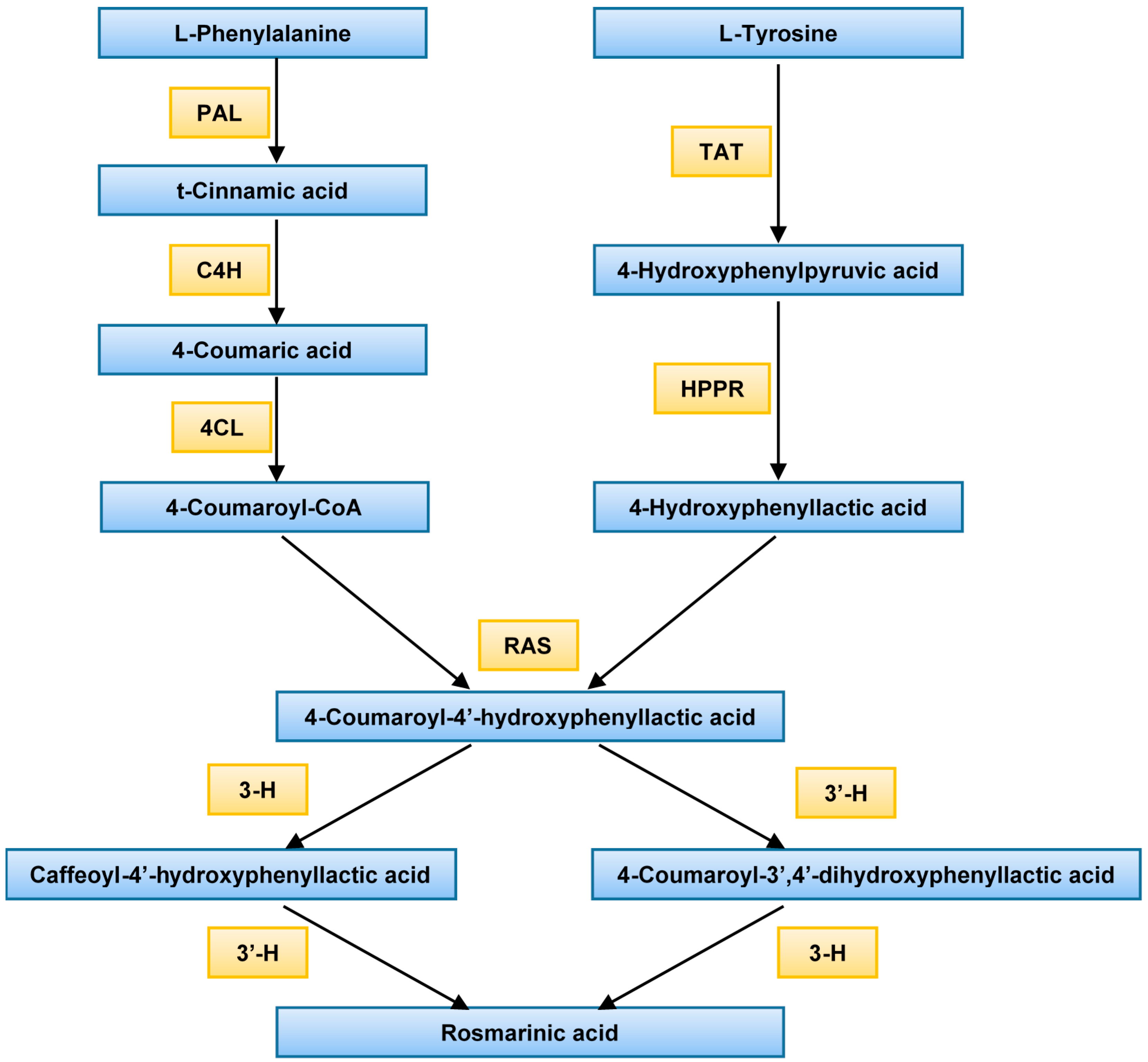
:1. Introduction
2. Results
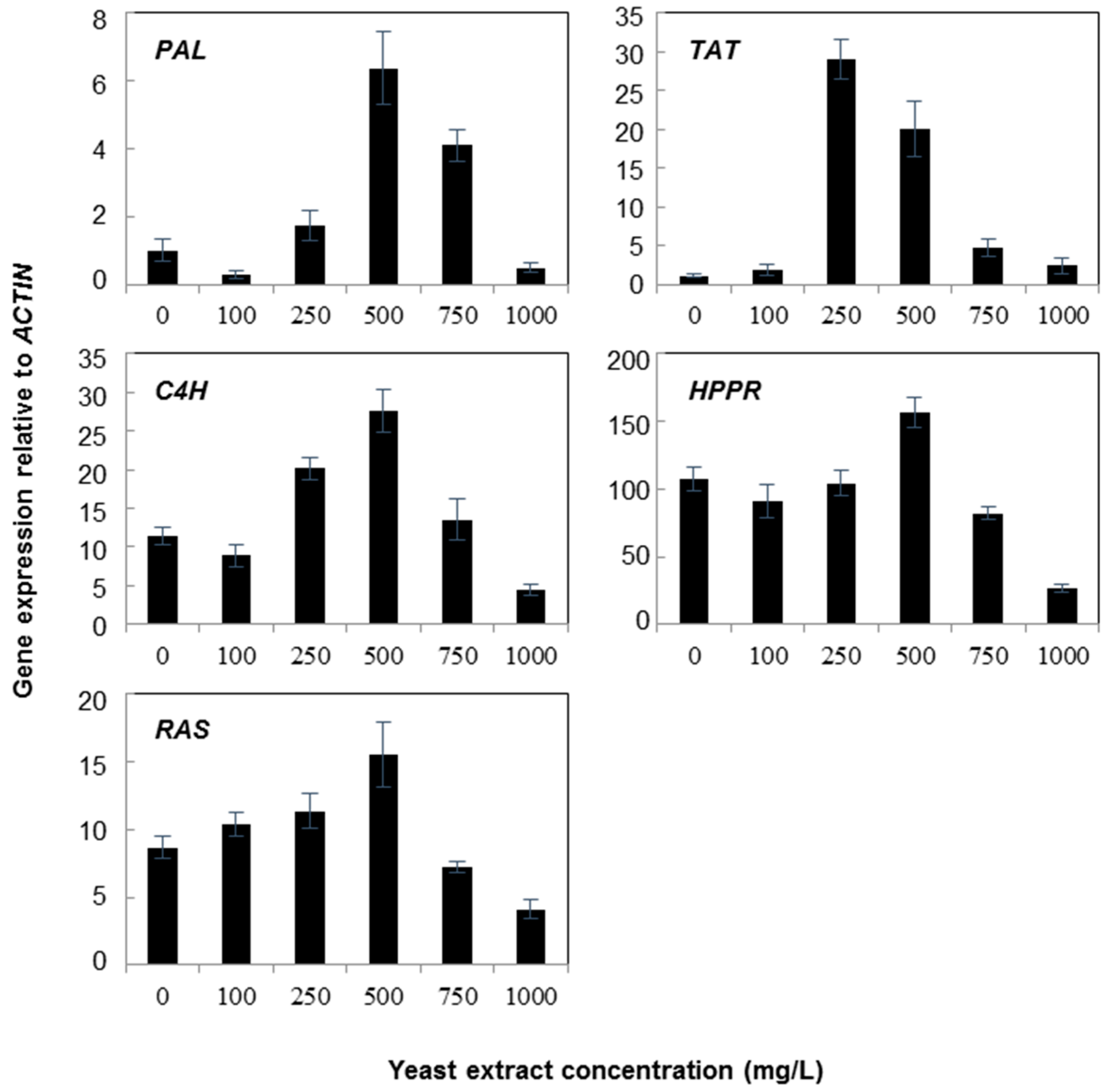
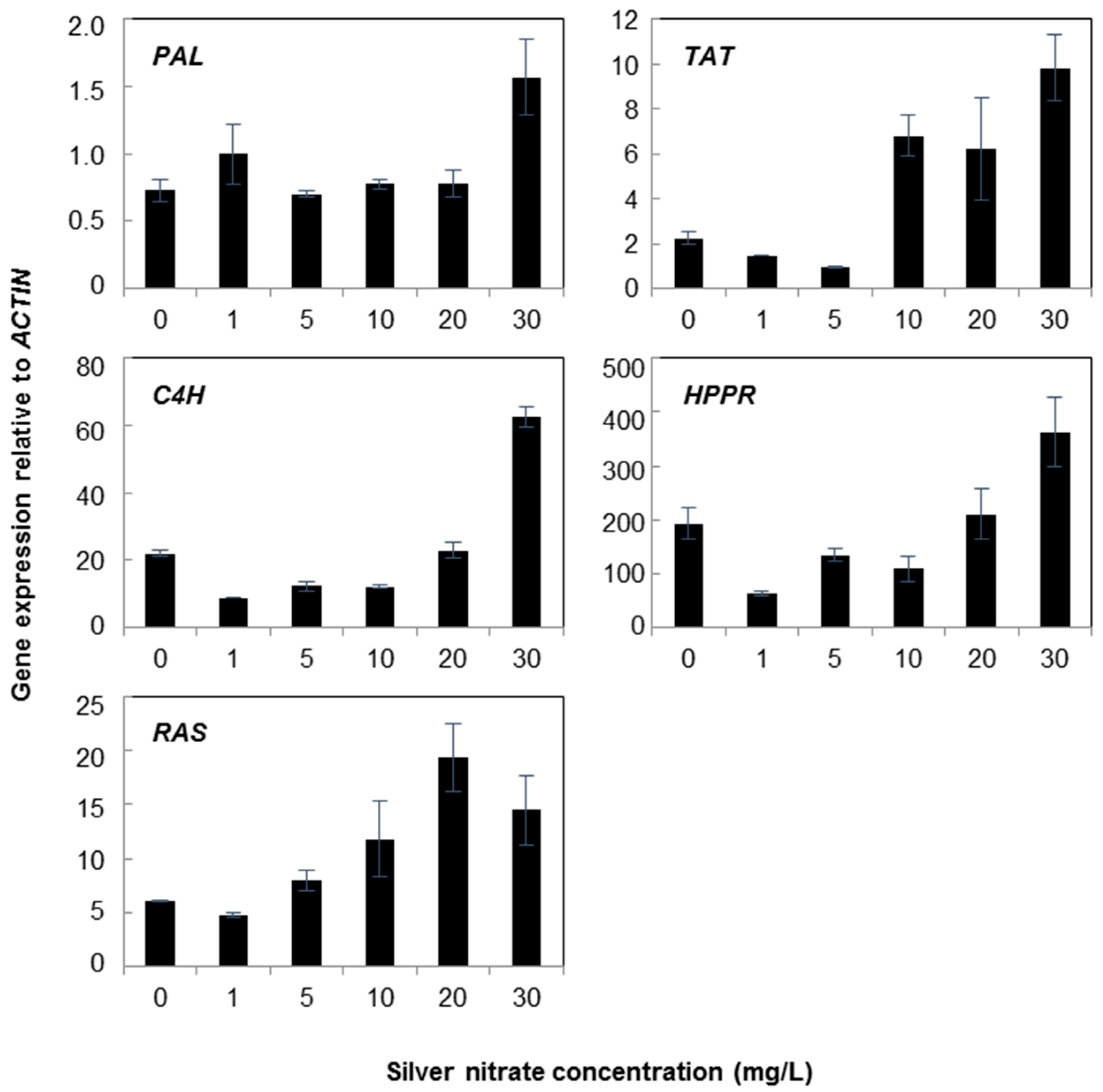
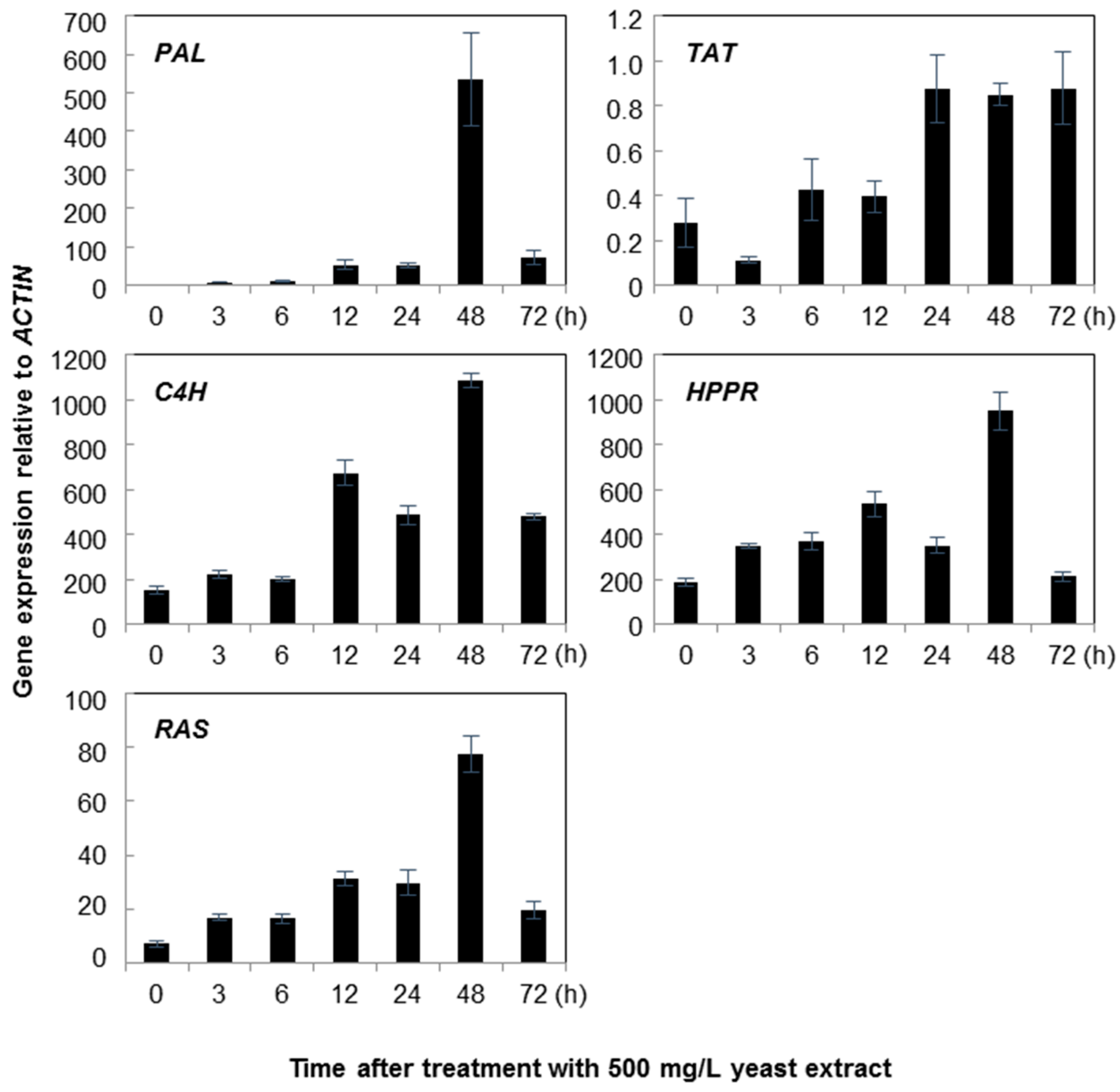
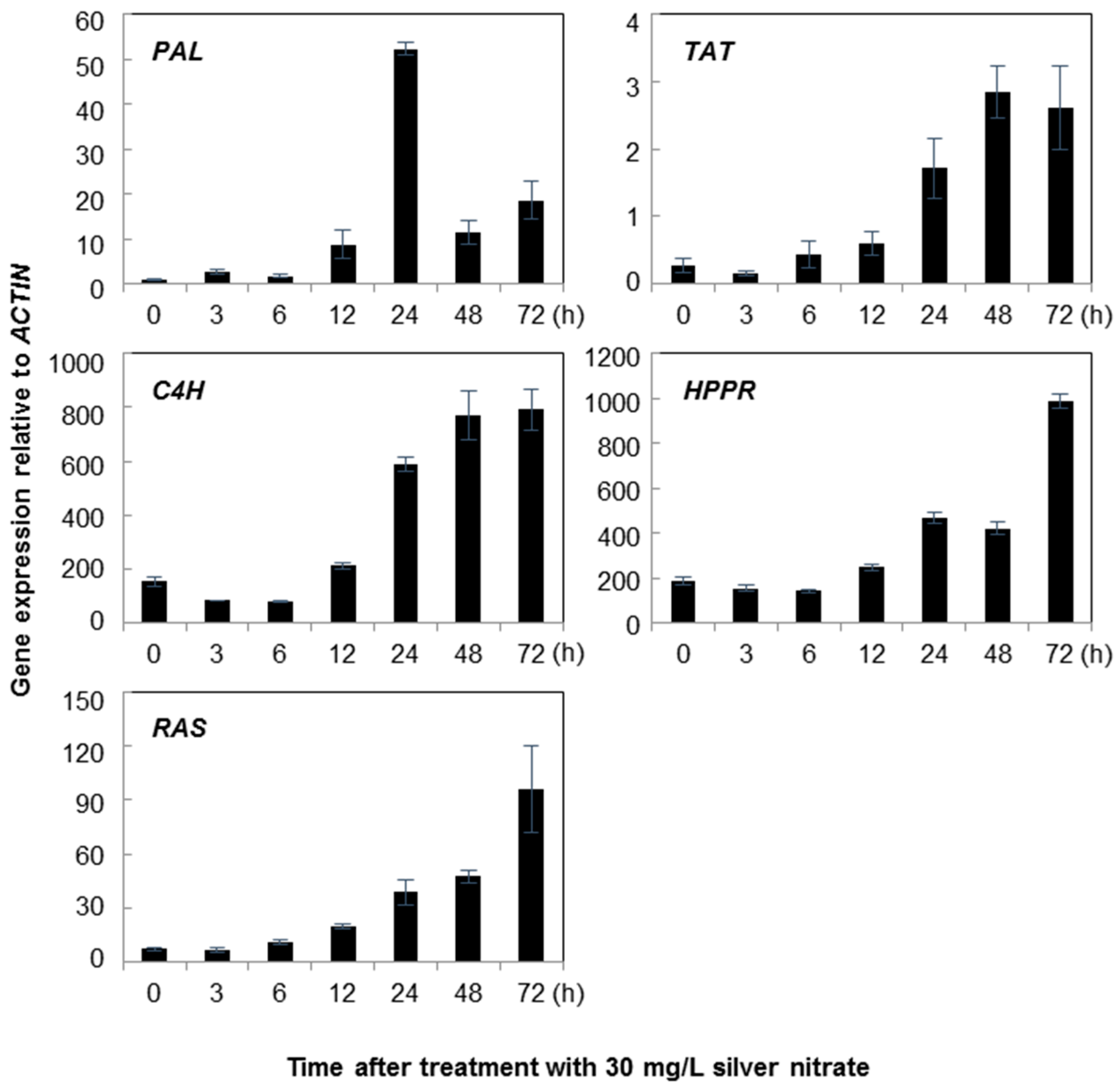
2.1. Gene Expression
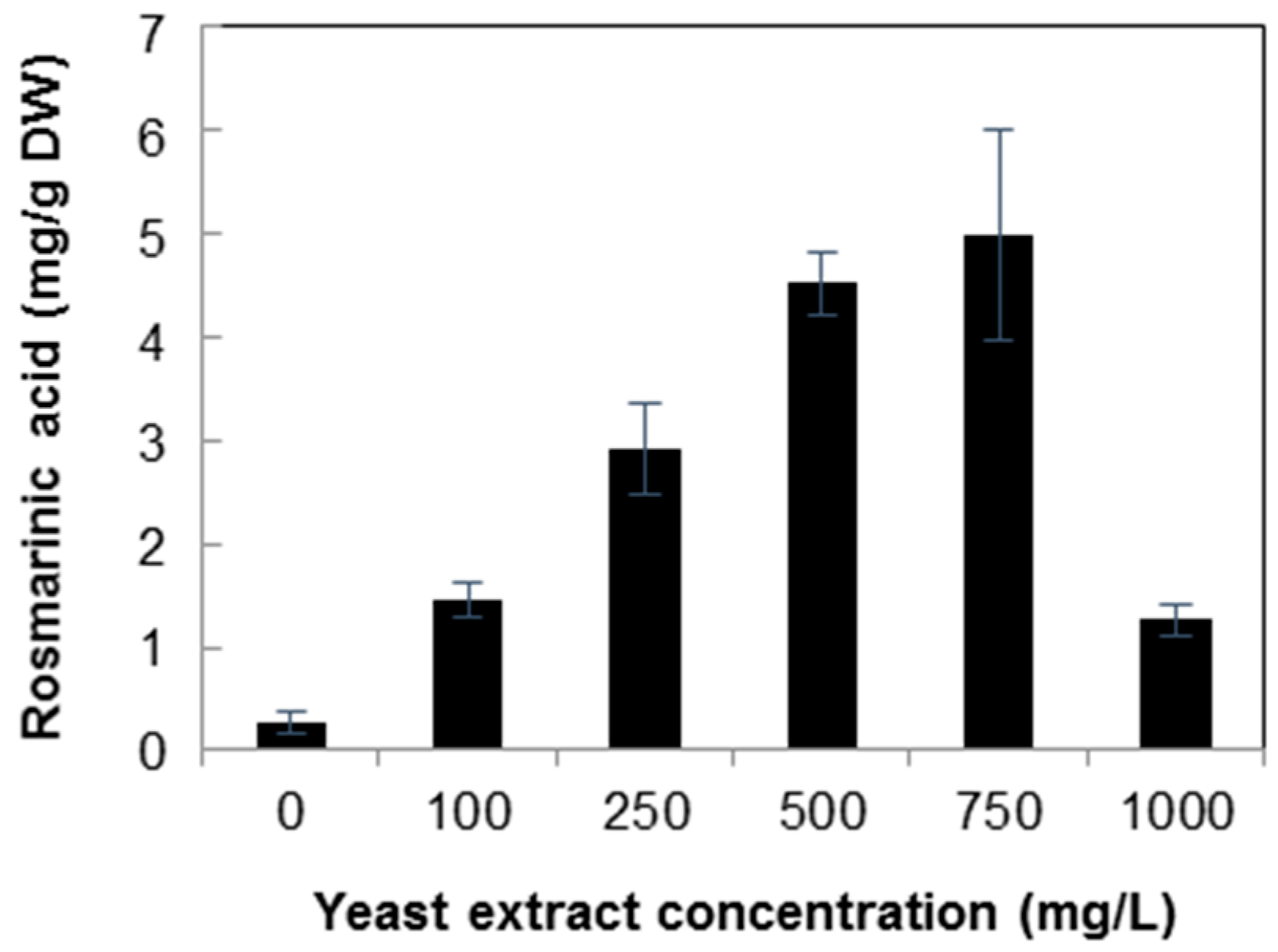
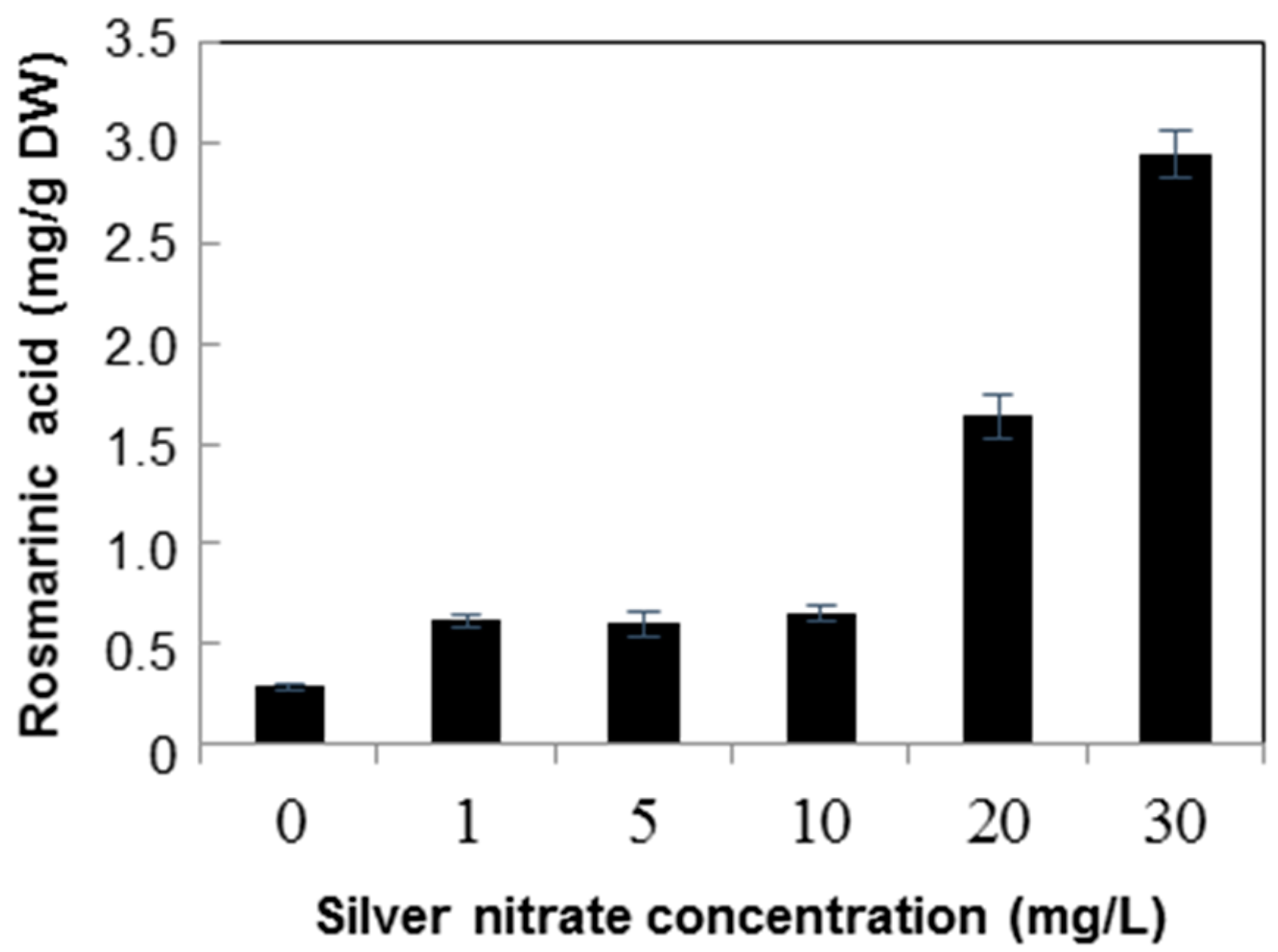
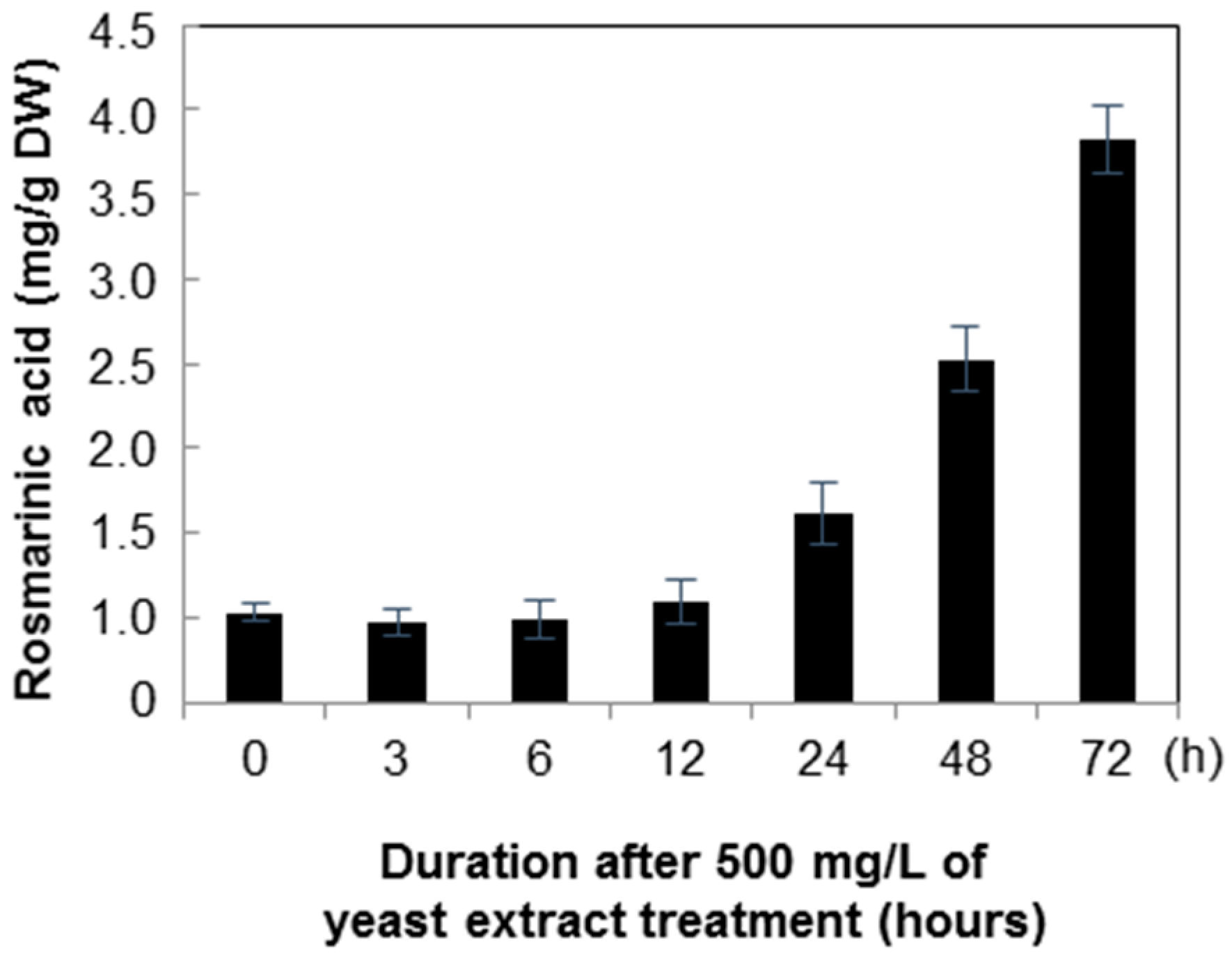
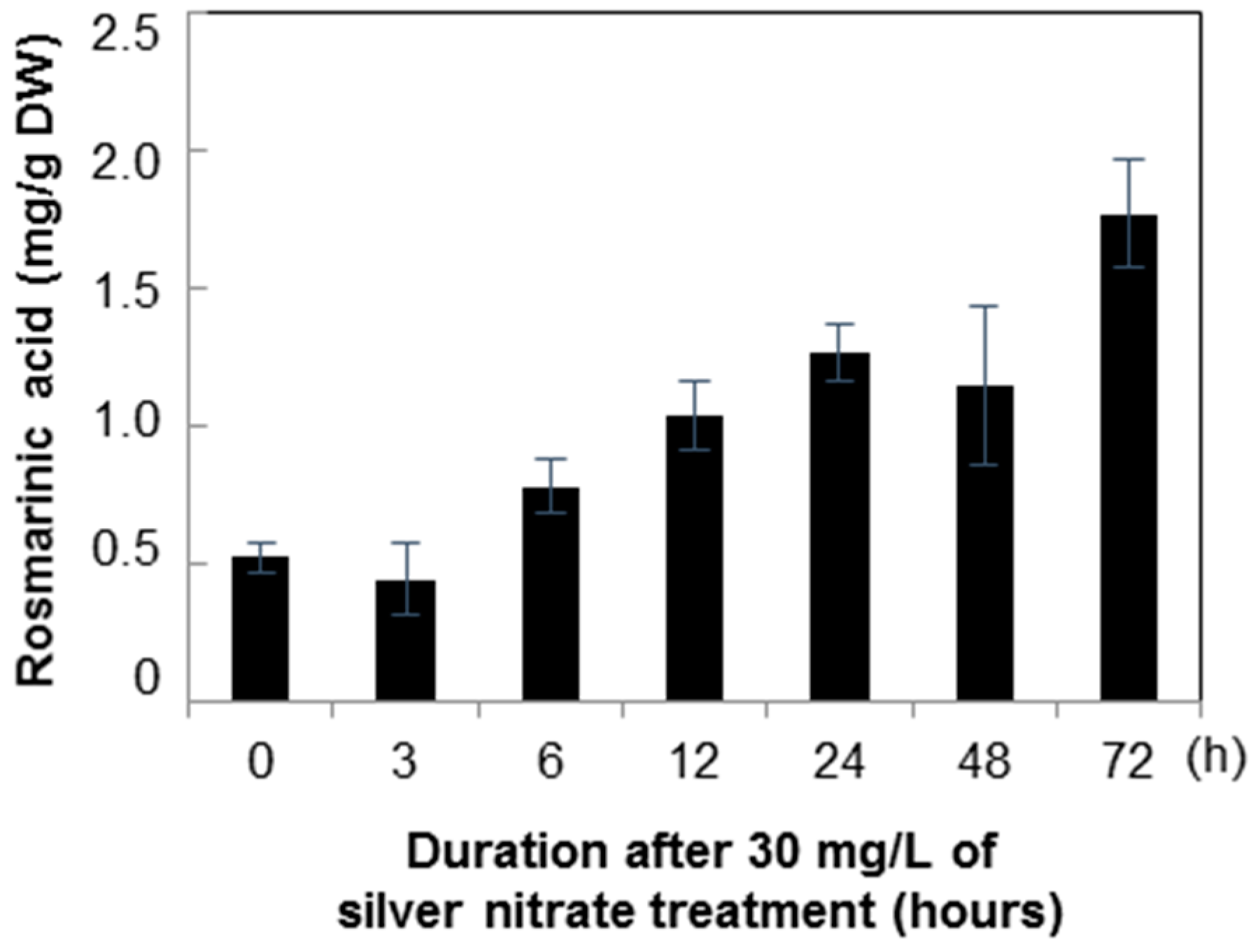
2.2. Quantification of Rosmarinic Acid
3. Discussion
4. Materials and Methods
4.1. Seed Germination and Callus Induction
4.2. Suspension Culture and Elicitor
4.3. Total RNA Extraction and cDNA Preparation
4.4. Quantitative Real Time-PCR for Gene Expression Analysis
4.5. Analysis of Phenylpropanoids by HPLC
4.6. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| PAL | Phenylalanine ammonia-lyase |
| C4H | Cinnamate 4-hydroxylase |
| 4CL | 4-Coumarate CoA ligase |
| TAT | Tyrosine amino transferase |
| HPPR | Hydroxyl phenylpyruvate reductase |
| RAS | Rosmarinic acid synthase (hydroxycinnamoyl-CoA:hydroxyphenyllactate hydroxycinnamoyl transferase) |
| 3-H | 3-Hydroxylase |
| 3′-H | 3′-Hydroxylase |
References
- Tepe, B.; Sokmen, A. Production and optimisation of rosmarinic acid by Satureja hortensis L. callus cultures. Nat. Prod. Res. 2007, 21, 1133–1144. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Shen, X.; Jain, R.; Lin, Y.; Wang, J.; Sun, J.; Wang, J.; Yan, Y.; Yuan, Q. Synthesis of chemicals by metabolic engineering of microbes. Chem. Soc. Rev. 2015, 44, 3760–3785. [Google Scholar] [CrossRef] [PubMed]
- Dornenburg, H.; Knorr, D. Strategies for the improvement of secondary metabolite production in plant cell cultures. Enzym. Microb. Technol. 1995, 17, 674–684. [Google Scholar] [CrossRef]
- Zhong, J.J. Biochemical engineering of the production of plant specific secondary metabolites by cell suspension cultures. In Advances in Biochemical Engineering/Biotechnology; Springer: Berlin/Heidelberg, Germany, 2001; pp. 1–26. [Google Scholar]
- Smetanska, I. Production of secondary metabolites using plant cell cultures. Food Biotechnol. 2008, 111, 187–228. [Google Scholar]
- Discosmo, F.; Misawa, M. Eliciting secondary metabolism in plant cell cultures. Trends Biotechnol. 1985, 3, 318–322. [Google Scholar] [CrossRef]
- Tamari, G.; Borochov, A.; Atzorn, R.; Weiss, D. Methyl jasmonate induces pigmentation and flavonoid gene expression in Petunia corollas: A possible role in wound response. Physiol. Plant. 1995, 94, 45–50. [Google Scholar] [CrossRef]
- Tyler, R.T.; Eilert, U.; Rijnders, C.O.M.; Roewer, I.A.; McNabb, C.K.; Kurz, W.G.W. Studies on Benzophenanthridine alkaloid production in elicited cell cultures of Papaver somniferum L. In Primary and Secondary Metabolism of Plant Cell Cultures II; Springer: Berlin/Heidelberg, Germany, 1989; pp. 200–207. [Google Scholar]
- Bostock, R.M.; Laine, R.A.; Kuć, J.A. Factors affecting the elicitation of sesquiterpenoid phytoalexin accumulation by eicosapentaenoic and arachidonic acids in potato. Plant Physiol. 1982, 70, 1417–1424. [Google Scholar] [CrossRef] [PubMed]
- Conrath, U.; Domard, A.; Kauss, H. Chitosan-elicited synthesis of callose and of coumarin derivatives in parsley cell suspension cultures. Plant Cell Rep. 1989, 8, 152–155. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.J.; Choi, J.H.; Oh, S.R.; Lee, H.K.; Park, J.H.; Lee, K.Y.; Kim, J.J.; Jeong, T.S.; Oh, G.T. Inhibition of cytokine-induced vascular cell adhesion molecule-1 expression; possible mechanism for anti-atherogenic effect of Agastache rugosa. FEBS Lett. 2001, 495, 142–147. [Google Scholar] [CrossRef]
- Song, J.H.; Kim, M.J.; Kwon, H.D.; Park, I.H. Antimicrobial activity and components of extracts from Agastache rugosa during growth period. J. Food Sci. Nutr. 2001, 6, 10–15. [Google Scholar]
- Shin, S.; Kang, C.A. Antifungal activity of the essential oil of Agastache rugosa Kuntze and its synergism with ketoconazole. Lett. Appl. Microbiol. 2003, 36, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.P.; Wei, H.L.; Zhao, H.S.; Xiao, S.Y.; Zheng, R.L. Antiapoptotic and antioxidant effects of rosmarinic acid in astrocytes. Pharmazie 2005, 60, 62–65. [Google Scholar] [PubMed]
- Nam, K.H.; Choi, J.H.; Seo, Y.J.; Lee, Y.M.; Won, Y.S.; Lee, M.R.; Lee, M.N.; Park, J.G.; Kim, Y.M.; Kim, H.C. Inhibitory effects of tilianin on the expression of inducible nitric oxide synthase in low density lipoprotein receptor deficiency mice. Exp. Mol. Med. 2006, 38, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Swarup, V.; Ghosh, J.; Ghosh, S.; Saxena, A.; Basu, A. Antiviral and anti-inflammatory effects of rosmarinic acid in an experimental murine model of Japanese encephalitis. Antimicrob. Agents Chemother. 2007, 51, 3367–3370. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Abreu, O.; Castillo-España, P.; León-Rivera, I.; Ibarra-Barajas, M.; Villalobos-Molina, R.; González-Christen, J.; Vergara-Galicia, J.; Estrada-Soto, S. Antihypertensive and vasorelaxant effects of tilianin isolated from Agastache Mexicana are mediated by NO/cGMP pathway and potassium channel opening. Biochem. Pharmacol. 2009, 78, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Al-Dhabi, N.A.; Arasu, M.V.; Park, C.H.; Park, S.U. Recent studies on rosmarinic acid and its biological and pharmacological activities. Excli J. 2014, 13, 1192–1195. [Google Scholar] [PubMed]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M.S. Characterization of rosmarinic acid synthase from cell cultures of Coleus blumei. Phytochemistry 1991, 30, 2877–2881. [Google Scholar] [CrossRef]
- Petersen, M. Cytochrome P450-dependent hydroxylation in the biosynthesis of rosmarinic acid in Coleus. Phytochemistry 1997, 45, 1165–1172. [Google Scholar] [CrossRef]
- Zenk, M.; El-Shagi, H.; Ulbrich, B. Production of rosmarinic acid by cell-suspension cultures of Coleus blumei. Naturwissenschaften 1977, 64, 585–586. [Google Scholar] [CrossRef]
- Razzaque, A.; Ellis, B. Rosmarinic acid production in coleus cell cultures. Planta 1977, 137, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Szabo, E.; Thelen, A.; Petersen, M. Fungal elicitor preparations and methyl jasmonate enhance rosmarinic acid accumulation in suspension cultures of Coleus blumei. Plant Cell Rep. 1999, 18, 485–489. [Google Scholar] [CrossRef]
- Xu, H.; Kim, Y.K.; Jin, X.; Lee, S.Y.; Park, S.U. Rosmarinic acid biosynthesis in callus and cell cultures of Agastache rugosa Kuntze. J. Med. Plants Res. 2008, 2, 237–241. [Google Scholar]
- Kintzios, S.; Makri, O.; Panagiotopoulos, E.; Scapeti, M. In vitro rosmarinic acid accumulation in sweet basil (Ocimum basilicum L.). Biotechnol. Lett. 2003, 25, 405–408. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Oh, S.R.; Lee, H.K.; Huh, H. Benzothiadiazole enhances the elicitation of rosmarinic acid production in a suspension culture of Agastache rugosa O. Kuntze. Biotechnol. Lett. 2001, 23, 55–60. [Google Scholar] [CrossRef]
- Mizukami, H.; Ogawa, T.; Ohashi, H.; Ellis, B.E. Induction of rosmarinic acid biosynthesis in Lithospermum erythrorhizon cell suspension cultures by yeast extract. Plant Cell Rep. 1992, 11, 480–483. [Google Scholar] [CrossRef] [PubMed]
- Sumaryono, W.; Proksch, P.; Hartmann, T.; Nimtz, M.; Wray, V. Induction of rosmarinic acid accumulation in cell suspension cultures of Orthosiphon aristatus after treatment with yeast extract. Phytochemistry 1991, 30, 3267–3271. [Google Scholar] [CrossRef]
- Yan, Q.; Shi, M.; Ng, J.; Wu, J.Y. Elicitor-induced rosmarinic acid accumulation and secondary metabolism enzyme activities in Salvia miltiorrhiza hairy roots. Plant Sci. 2006, 170, 853–858. [Google Scholar] [CrossRef]
- Mizukami, H.; Tabira, Y.; Ellis, B.E. Methyl jasmonate-induced rosmarinic acid biosynthesis in Lithospermum erythrorhizon cell suspension cultures. Plant Cell Rep. 1993, 12, 706–709. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Arnaldos, T.; Lopez-Serrano, M.; Ros, B.A.; Calderon, A.; Zapata, J. Tentative evidence of a rosmarinic acid peroxidase in cell cultures from lavandin (Lavandula x intermedia) flowers. Biochem. Mol. Biol. Int. 1994, 34, 809–816. [Google Scholar] [PubMed]
- Kračun-Kolarević, M.; Dmitrović, S.; Filipović, B.; Perić, M.; Mišić, D.; Simonović, A.; Todorović, S. Influence of sodium salicylate on rosmarinic acid, carnosol and carnosic acid accumulation by Salvia officinalis L. shoots grown in vitro. Biotechnol. Lett. 2015, 37, 1693–1701. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Xu, H.; Kim, Y.; Park, S. Rosmarinic acid production in hairy root cultures of Agastache rugosa Kuntze. World J. Microbiol. Biotechnol. 2008, 24, 969–972. [Google Scholar]
- Park, S.U.; Uddin, M.R.; Xu, H.; Kim, Y.K.; Lee, S.Y. Biotechnological applications for rosmarinic acid production in plant. Afr. J. Biotechnol. 2008, 7, 4959–4965. [Google Scholar]
- Kim, Y.B.; Kim, J.K.; Uddin, M.R.; Xu, H.; Park, W.T.; Tuan, P.A.; Li, X.; Chung, E.; Lee, J.H.; Park, S.U. Metabolomics analysis and biosynthesis of rosmarinic acid in Agastache rugosa Kuntze treated with methyl jasmonate. PLoS ONE 2013, 8, e64199. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds (rosmarinic acid and tilianin) are available from the authors.
| Primers | Sequences (5′ to 3′) | Amplicon Size (bp) |
|---|---|---|
| ArActin F | ACCTCAAAATAGCATGGGGAAGT | 151 |
| ArActin R | GGCCGTTCTCTCACTTTATGCTA | |
| ArPAL F | ACGGCTCCAACGGTCATAATAAT | 108 |
| ArPAL R | ATCCGCTTTACCTCCTCAAGGT | |
| ArC4H F | GTTCGAGAGTGAGAATGATCCGT | 157 |
| ArC4H R | ATAATCCTTGAACAATTGCAGCC | |
| ArHPPR F | AAGGGGATTAGGGTTACCAACACG | 200 |
| ArHPPR R | ATTCTGCCCAATCCTATGATGCC | |
| ArTAT F | AGGCAGCAGTACCAGCCATTCTT | 163 |
| ArTAT R | TTGACCATGAAAGCCATTGATCC | |
| ArRAS F | GGCGAACTACCACACGCTGAG | 161 |
| ArRAS R | CGATCTCGAGACGGTTATTGTCG |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, W.T.; Arasu, M.V.; Al-Dhabi, N.A.; Yeo, S.K.; Jeon, J.; Park, J.S.; Lee, S.Y.; Park, S.U. Yeast Extract and Silver Nitrate Induce the Expression of Phenylpropanoid Biosynthetic Genes and Induce the Accumulation of Rosmarinic Acid in Agastache rugosa Cell Culture. Molecules 2016, 21, 426. https://doi.org/10.3390/molecules21040426
Park WT, Arasu MV, Al-Dhabi NA, Yeo SK, Jeon J, Park JS, Lee SY, Park SU. Yeast Extract and Silver Nitrate Induce the Expression of Phenylpropanoid Biosynthetic Genes and Induce the Accumulation of Rosmarinic Acid in Agastache rugosa Cell Culture. Molecules. 2016; 21(4):426. https://doi.org/10.3390/molecules21040426
Chicago/Turabian StylePark, Woo Tae, Mariadhas Valan Arasu, Naif Abdullah Al-Dhabi, Sun Kyung Yeo, Jin Jeon, Jong Seok Park, Sook Young Lee, and Sang Un Park. 2016. "Yeast Extract and Silver Nitrate Induce the Expression of Phenylpropanoid Biosynthetic Genes and Induce the Accumulation of Rosmarinic Acid in Agastache rugosa Cell Culture" Molecules 21, no. 4: 426. https://doi.org/10.3390/molecules21040426






