In Silico Discovery of Potential Uridine-Cytidine Kinase 2 Inhibitors from the Rhizome of Alpinia mutica
Abstract
:1. Introduction
2. Results and Discussion
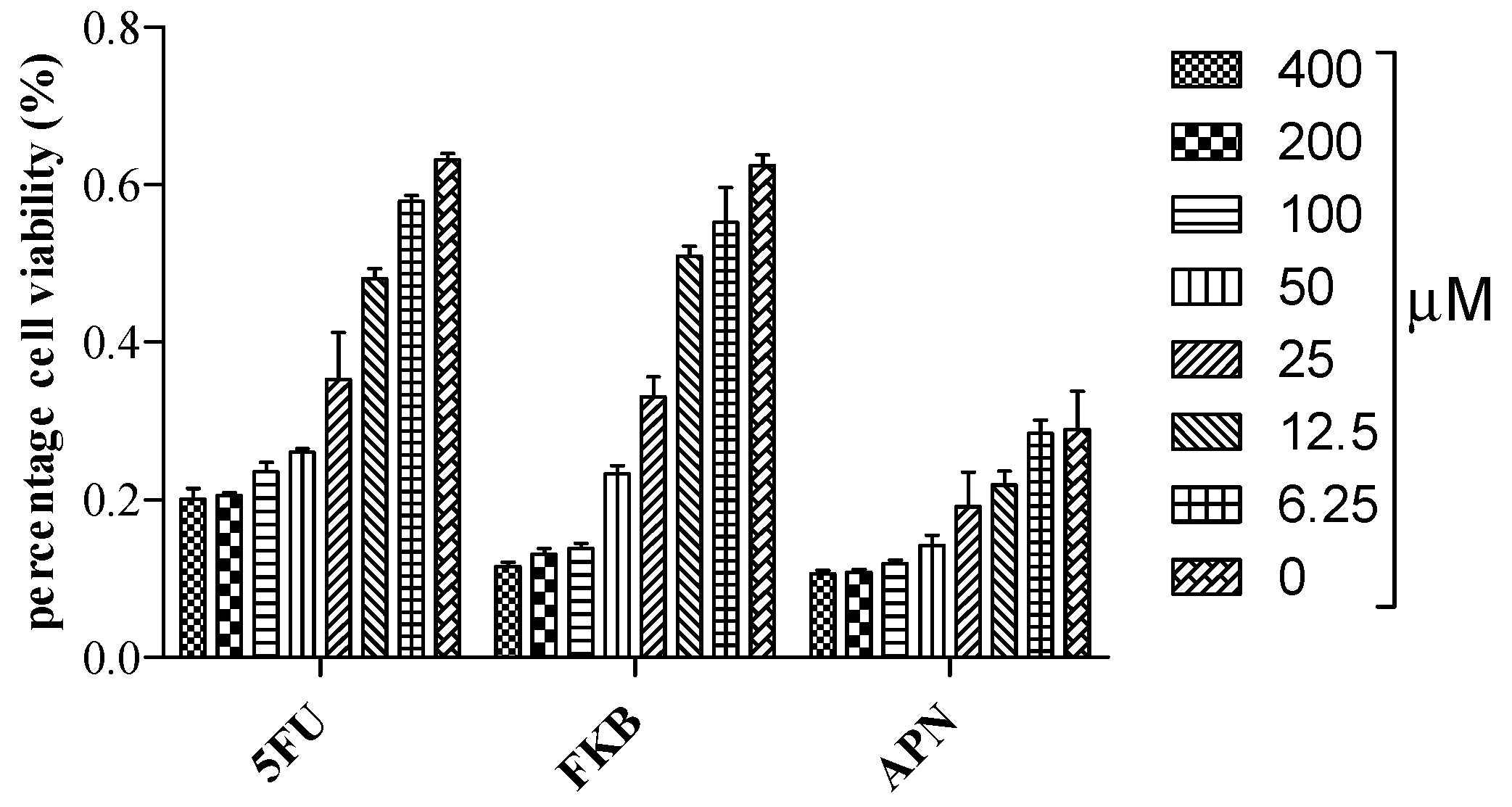
2.1. Cell Viability Studies
2.2. Molecular Docking Studies
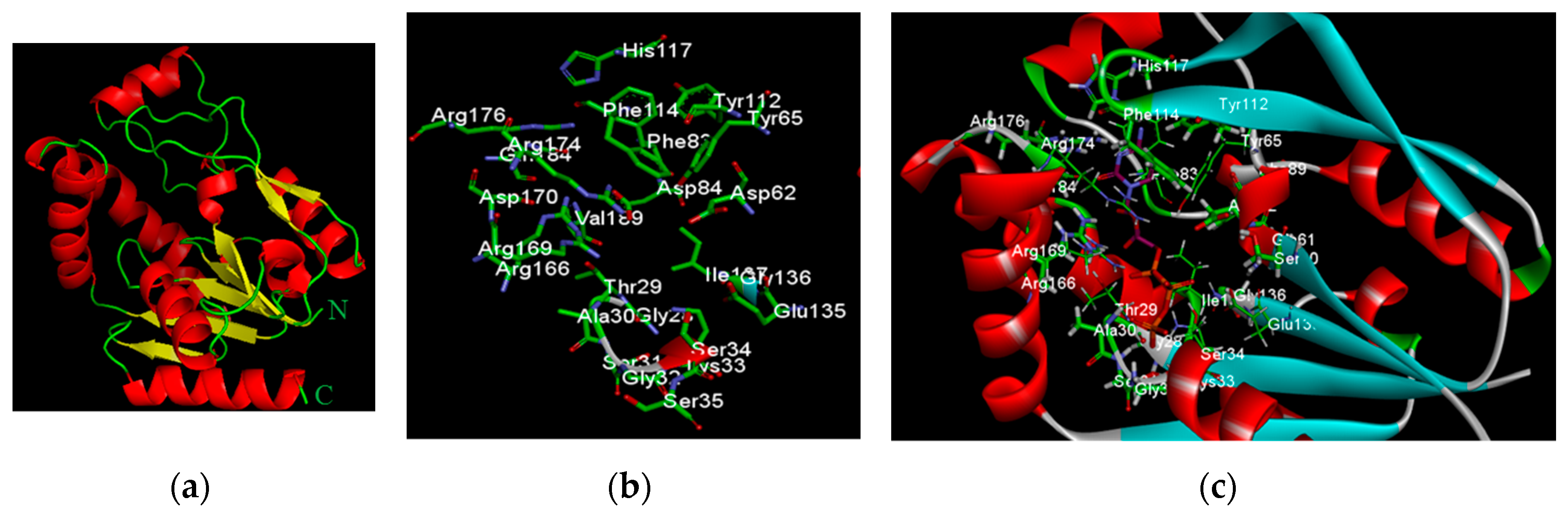
2.2.1. Redocking Analysis
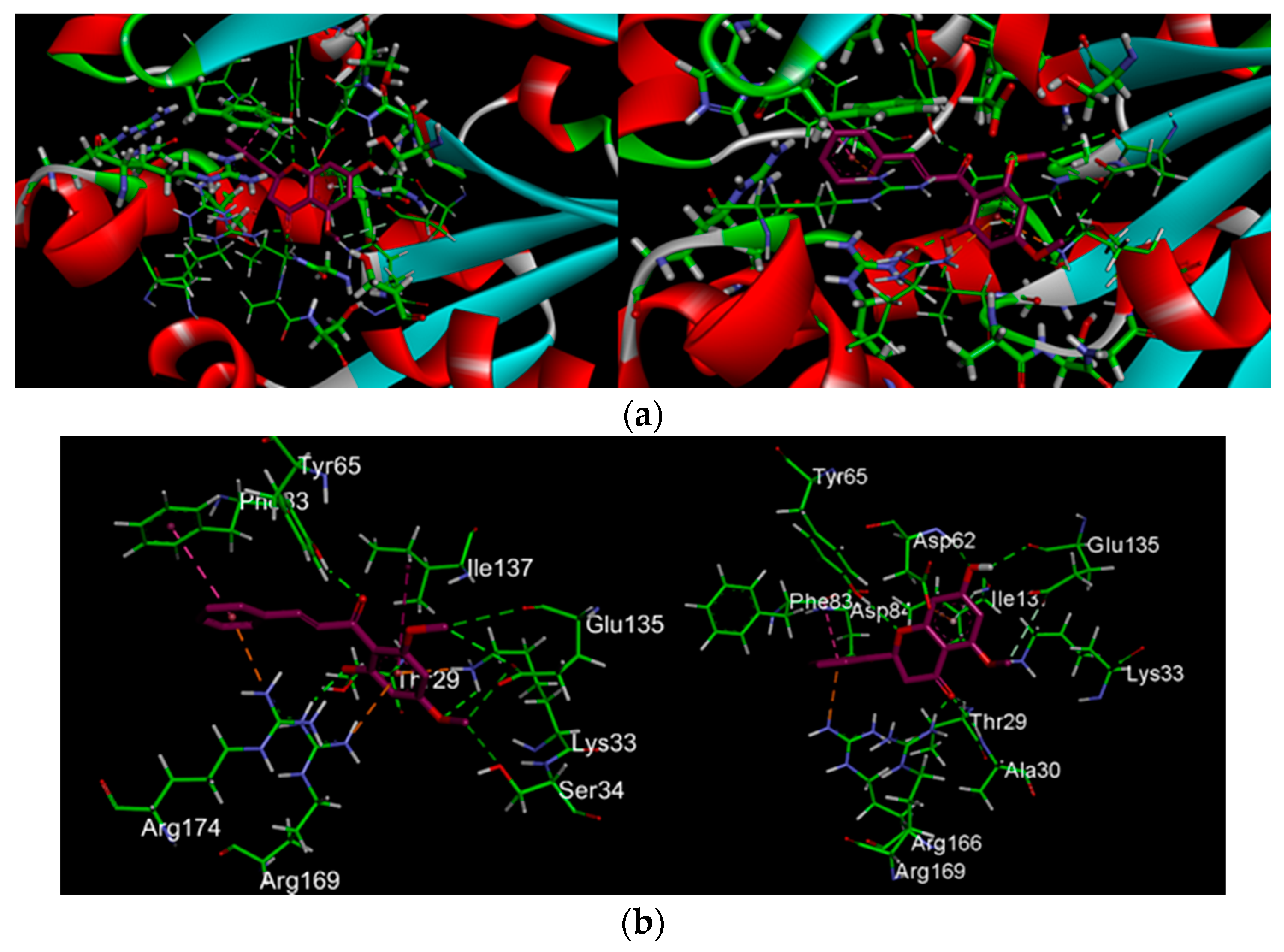
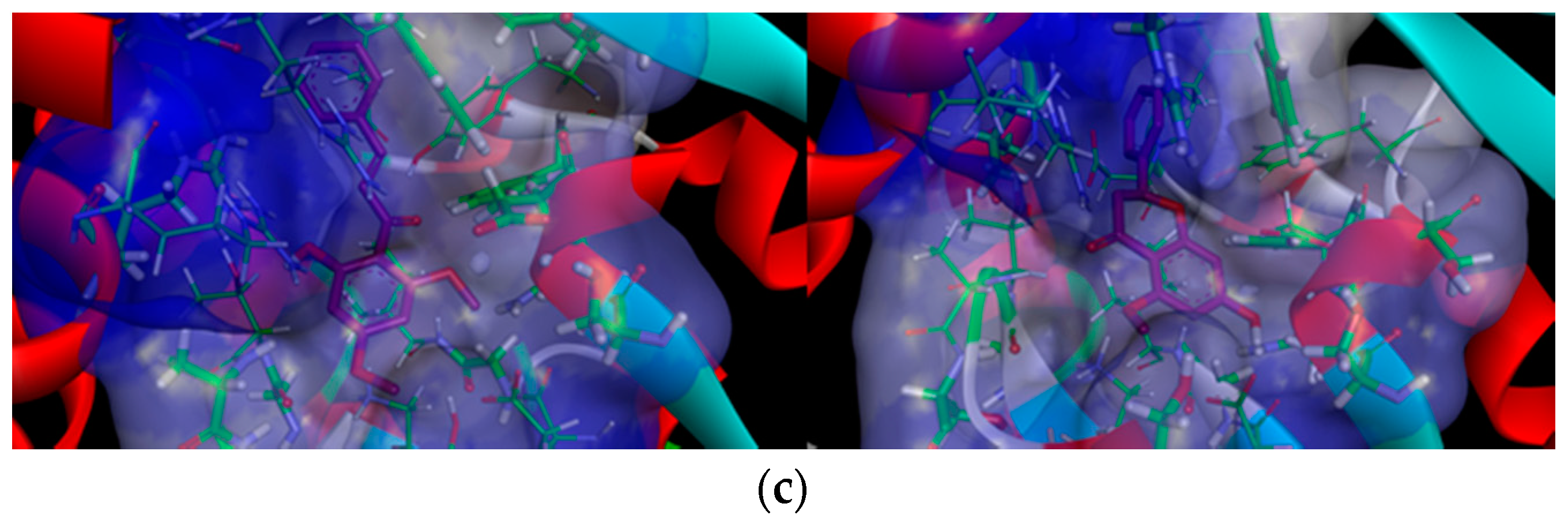
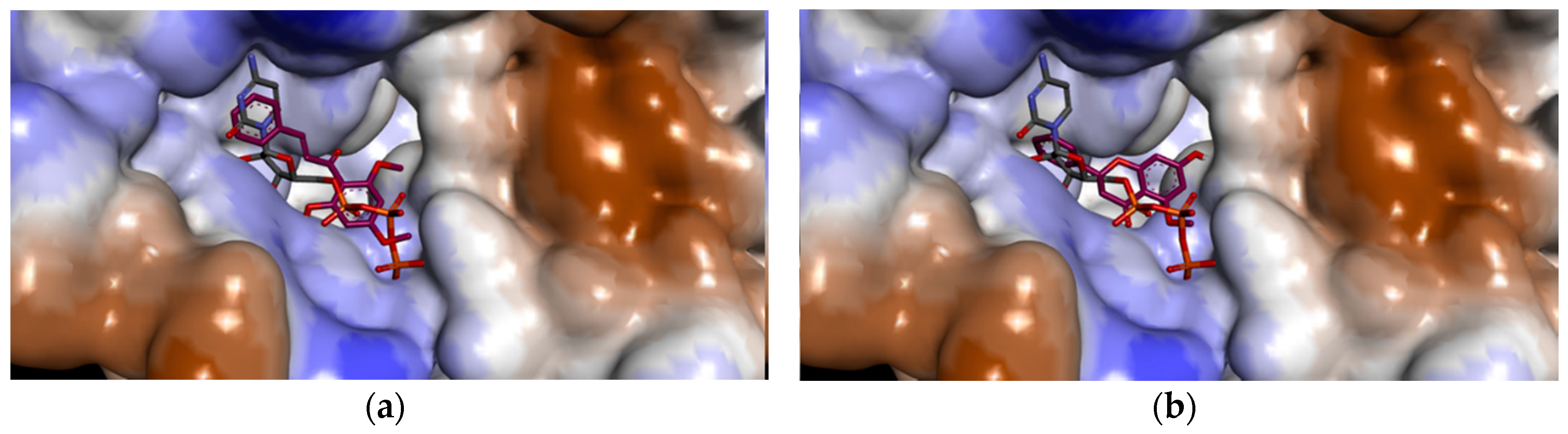
2.2.2. Docking Analysis of FKB and APN on UCK2 Protein
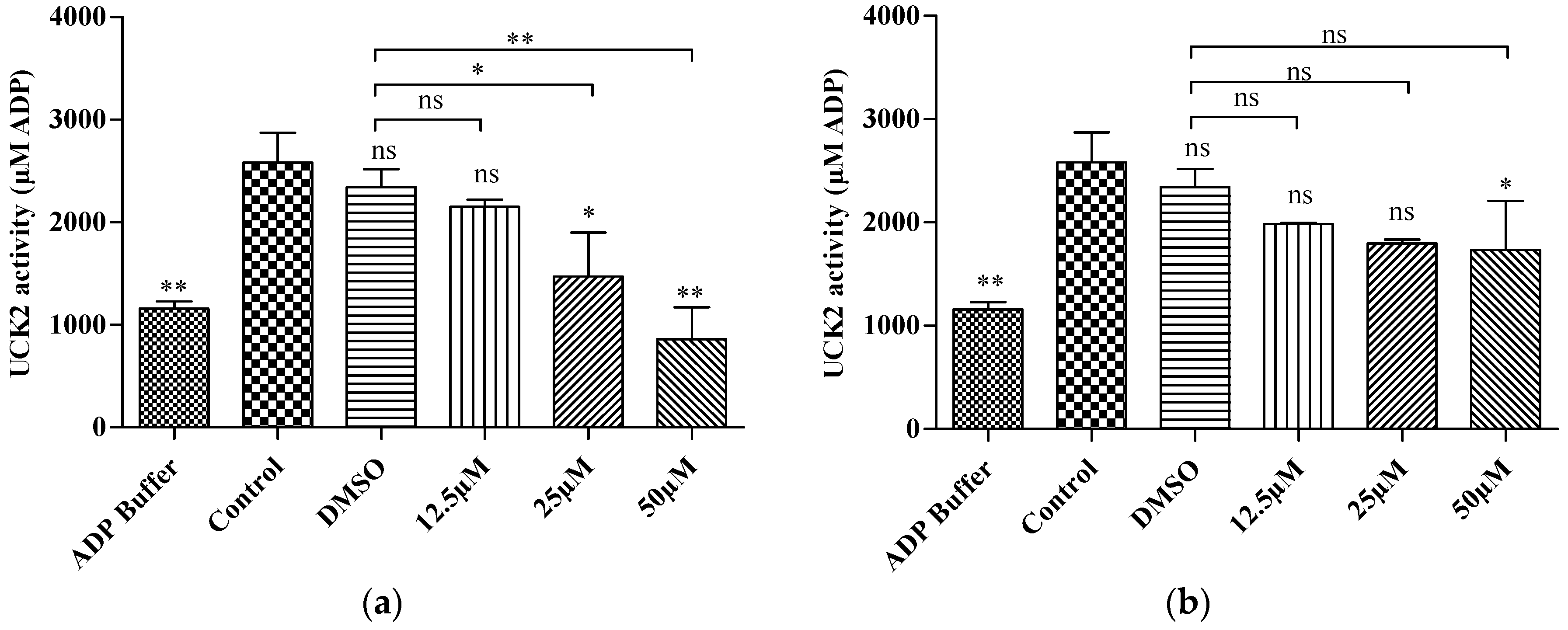
2.3. In Vitro Kinase Activity
3. Materials and Methods
3.1. General Information
3.2. Plant Material
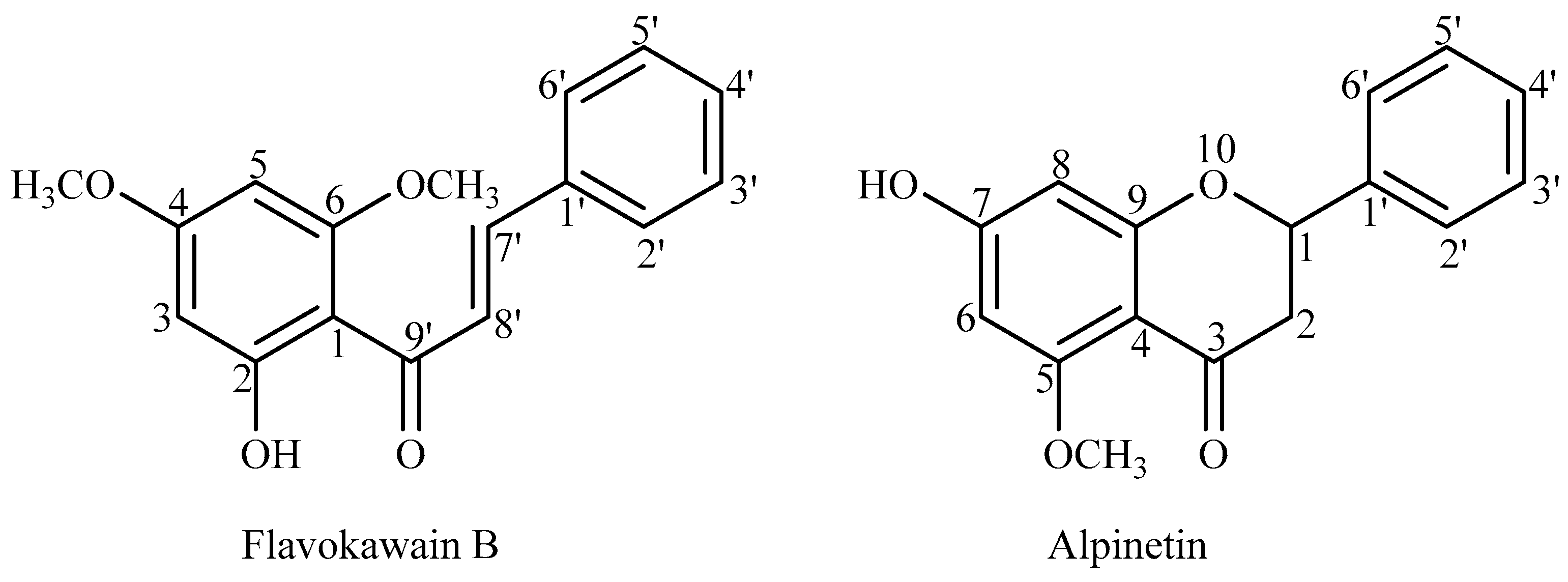
3.3. Extraction and Isolation of FKB and APN
3.4. Cell Culture
3.5. Drugs
3.6. Cell Viability Assay
3.7. Molecular Docking Simulation Studies
3.8. Preparation of Cell Lysate
3.9. In Vitro Kinase Activity Assay
3.10. Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wang, F.; Liu, X.; Liu, C.; Liu, Z.; Sun, L. Effects of antibiotic antitumor drugs on nucleotide levels in cultured tumor cells: An exploratory method to distinguish the mechanisms of antitumor drug action based on targeted metabolomics. Acta Pharm. Sin. B 2015, 5, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.M.; Tymoczko, J.L.; Stryer, L. Biochemistry, 5th ed.; W H Freeman: New York, NY, USA, 2002. [Google Scholar]
- Zhang, C.; Liu, Z.; Liu, X.; Wei, L.; Liu, Y.; Yu, J.; Sun, L. Targeted metabolic analysis of nucleotides and identification of biomarkers associated with cancer in cultured cell models. Acta Pharm. Sin. B 2013, 3, 254–262. [Google Scholar] [CrossRef]
- Lane, A.N.; Fan, T.W.-M. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res. 2015, 43, 2466–2485. [Google Scholar] [CrossRef] [PubMed]
- Connolly, G.P.; Duley, J.A. Uridine and its nucleotides: Biological actions, therapeutic potentials. Trends Pharmacol. Sci. 1999, 20, 218–225. [Google Scholar] [CrossRef]
- Van Rompay, A.R.; Norda, A.; Lindén, K.; Johansson, M.; Karlsson, A. Phosphorylation of uridine and cytidine nucleoside analogs by two human uridine-cytidine kinases. Mol. Pharmacol. 2001, 59, 1181–1186. [Google Scholar] [PubMed]
- Appleby, T.C.; Larson, G.; Cheney, I.W.; Walker, H.; Wu, J.Z.; Zhong, W.; Hong, Z.; Yao, N. Structure of human uridine-cytidine kinase 2 determined by SIRAS using a rotating-anode X-ray generator and a single samarium derivative. Acta Crystallogr. D Biol. Crystallogr. 2005, 61, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Friday, B.; Lassere, Y.; Meyers, C.A.; Mita, A.; Abbruzzese, J.L.; Thomas, M.B. A phase I study to determine the safety and pharmacokinetics of intravenous administration of TAS-106 once per week for three consecutive weeks every 28 days in patients with solid tumors. Anticancer Res. 2012, 32, 1689–1696. [Google Scholar] [PubMed]
- Naing, A.; Fu, S.; Zinner, R.G.; Wheler, J.J.; Hong, D.S.; Arakawa, K.; Falchook, G.S.; Kurzrock, R. Phase I dose-escalating study of TAS-106 in combination with carboplatin in patients with solid tumors. Investig. New Drugs 2013, 32, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Peters, G.J.; Julsing, J.R.; Smid, K.; De Klerk, D.; Sarkisjan, D.; Yang, M.Y.; Lee, Y.B.; Kim, D.J. Fluorocyclopentenylcytosine (RX-3117) is activated by uridine-cytidine kinase 2, a potential biomarker. In Proceedings of the 106th Annual Meeting of the American Association for Cancer Research, Philadelphia, PA, USA, 18–22 April 2015.
- Hammond-Thelin, L.A.; Thomas, M.B.; Iwasaki, M.; Abbruzzese, J.L.; Lassere, Y.; Meyers, C.A; Hoff, P.; de Bono, J.; Norris, J.; Matsushita, H.; et al. Phase I and pharmacokinetic study of 3’-C-ethynylcytidine (TAS-106), an inhibitor of RNA polymerase I, II and III, in patients with advanced solid malignancies. Investig. New Drugs 2012, 30, 316–326. [Google Scholar] [CrossRef] [PubMed]
- Tsao, A.; Hui, E.P.; Juergens, R.; Marur, S.; Huat, T.E.; Cher, G.B.; Hong, R.-L.; Hong, W.K.; Chan, A.T.-C. Phase II study of TAS-106 in patients with platinum-failure recurrent or metastatic head and neck cancer and nasopharyngeal cancer. Cancer Med. 2013, 2, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Mayer, R.J.; Van Cutsem, E.; Falcone, A.; Yoshino, T.; Garcia-Carbonero, R.; Mizunuma, N.; Yamazaki, K.; Shimada, Y.; Tabernero, J.; Komatsu, Y.; et al. Randomized trial of TAS-102 for refractory metastatic colorectal cancer. N. Engl. J. Med. 2015, 372, 1909–1919. [Google Scholar] [CrossRef] [PubMed]
- Pandit, N.K. Introduction to the Pharmaceutical Sciences, 1st ed.; Lippincott Williams &Wilkins: Philadelphia, PA, USA, 2007. [Google Scholar]
- Kuo, Y.F.; Su, Y.Z.; Tseng, Y.H.; Wang, S.Y.; Wang, H.M.; Chueh, P.J. Flavokawain B, a novel chalcone from Alpinia pricei Hayata with potent apoptotic activity: Involvement of ROS and GADD153 upstream of mitochondria-dependent apoptosis in HCT116 cells. Free Radic. Biol. Med. 2010, 49, 214–226. [Google Scholar] [CrossRef] [PubMed]
- Malek, S.N.A.; Phang, C.W.; Ibrahim, H.; Wahab, N.A.; Sim, K.S. Phytochemical and cytotoxic investigations of Alpinia mutica rhizomes. Molecules 2011, 16, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.P.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.N.; Koizumi, K.; Fukushima, M.; Matsuda, A.; Inagaki, F. Structural basis for the specificity, catalysis, and regulation of human uridine-cytidine kinase. Structure 2004, 12, 751–764. [Google Scholar] [CrossRef] [PubMed]
- Glickman, J.F. Assay Development for Protein Kinase Enzymes. In Assay Guidance Manual; The Rockefeller University: New York, NY, USA, 2004; (available in NCBI: NBK53196). [Google Scholar]
- Mustahil, N.A.; Sukari, M.A.; Abdul, A.B.; Ali, N.A.; Ee, G.; Lian, C. Evaluation of biological activities of Alpinia mutica Roxb. and its chemical constituents. Pak. J. Pharm. Sci. 2013, 26, 391–395. [Google Scholar] [PubMed]
- Sirat, H.M.; Masri, D.; Rahm, A. Constituents of the Rhizomes of Alpinia rafflesiana. Pertanika J. Sci. Technol. 1996, 4, 17–22. [Google Scholar]
- Ranjith, W.; Dharmaratne, H.; Dhammika Nanayakkara, N.P.; Khan, I.A. Kavalactones from Piper methysticum, and their 13C NMR spectroscopic analyses. Phytochemistry 2002, 59, 429–433. [Google Scholar] [CrossRef]
- Yap, A.; Ching, L.; Wah, T.S.; Sukari, M.A.; Ee, G.; Lian, C. Characterization of flavonoid derivatives from Boesenbergia rotunda (L.). Malays. J. Anal. Sci. 2007, 11, 154–159. [Google Scholar]
- Zhou, P.; Gross, S.; Liu, J.-H.; Yu, B.-Y.; Feng, L.-L.; Nolta, J.; Sharma, V.; Piwnica-Worms, D.; Qiu, S.X. Flavokawain B, the hepatotoxic constituent from kava root, induces GSH-sensitive oxidative stress through modulation of IKK/NF-kappaB and MAPK signaling pathways. FASEB J. 2010, 24, 4722–4732. [Google Scholar]
- Xiao, X.; Si, X.; Tong, X.; Li, G. Preparation of flavonoids and diarylheptanoid from alpinia katsumadai hayata by microwave-assisted extraction and high-speed counter-current chromatography. Sep. Purif. Technol. 2011, 81, 265–269. [Google Scholar]
- PubChem. Available online: NCBI: http://www.ncbi.nlm.nih.gov/pccompound (accessed on 29 June 2015).
- Powers, C.N.; Setzer, W.N. A molecular docking study of phytochemical estrogen mimics from dietary herbal supplements. In Silico Pharmacol. 2015, 3, 4. [Google Scholar] [CrossRef]
- Saha, S.; Islam, M.; Shilpi, J.A.; Hasan, S. Inhibition of VEGF: A novel mechanism to control angiogenesis by Withania somnifera’s key metabolite Withaferin A. In Silico Pharmacol. 2013, 1, 11. [Google Scholar] [CrossRef]
- Sample Availability: Samples of flavokawain B and alpinetin are available from the authors.
| Ligand | Free Binding Energy * | Inhibition Constant, Ki | Intermolecular Energy * | vdW + Hbond + Desolv Energy * | Electrostatic Energy * |
|---|---|---|---|---|---|
| CTP | −14.27 | 34.63 pM | −16.66 | −12.59 | −4.07 |
| FKB | −8.47 | 618.12 nM | −10.26 | −9.96 | −0.30 |
| APN | −8.86 | 321.38 nM | −9.75 | −9.04 | −0.71 |
| UCK2 | HB with CTP | HB with FKB | HB with APN |
|---|---|---|---|
| Active Residues | (BD ≤ 3.5 Å, DHA ≥ 90°) | (BD ≤ 3.5 Å, DHA ≥ 90°) | (BD ≤ 3.5 Å, DHA ≥ 90°) |
| Thr29 | Hβ with 3′O | Oγ1 with 2 H | Hα with 3 O |
| Ala30 | Hα with Oγ3, HN with Oα2 | HN with 3 O | |
| Gly32 | HN with Oγ2 | ||
| Lys33 | Hζ2 with Oγ2, HN with Oγ2, Hζ1 with Oα1, Hζ1 with Oβ3 | Hζ2 with 4 O | Hζ1 with 5 O |
| Ser31 | HN with Oγ2 | ||
| Ser34 | HN with Oγ1, Hα with Oβ1, Hβ2 with Oγ1 | Oγ with 4′ H | |
| Asp62 | HN with 7 O | ||
| Tyr65 | Hη with 9′ O | Hη with 10 O | |
| Phe83 | |||
| Asp84 | Oγ1 with 2′O, Oγ2 with 3′O | ||
| Tyr112 | Oη with 4N | ||
| His117 | Nδ1 with 4N | ||
| Glu135 | Oε1 with 10′ H, 4′ H | Oε1 with 5 H | |
| O with 10′ H | O with 7 H | ||
| Arg166 | Oη12 with 3′O, Oη22 with 2′O, Oη22 with 3′O | ||
| Arg169 | Hη22 with Oγ2 | Hη22 with 2 O | Hη12 with 3O |
| Arg174 | Oη11 with 4′O | ||
| Arg179 |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malami, I.; Abdul, A.B.; Abdullah, R.; Bt Kassim, N.K.; Waziri, P.; Christopher Etti, I. In Silico Discovery of Potential Uridine-Cytidine Kinase 2 Inhibitors from the Rhizome of Alpinia mutica. Molecules 2016, 21, 417. https://doi.org/10.3390/molecules21040417
Malami I, Abdul AB, Abdullah R, Bt Kassim NK, Waziri P, Christopher Etti I. In Silico Discovery of Potential Uridine-Cytidine Kinase 2 Inhibitors from the Rhizome of Alpinia mutica. Molecules. 2016; 21(4):417. https://doi.org/10.3390/molecules21040417
Chicago/Turabian StyleMalami, Ibrahim, Ahmad Bustamam Abdul, Rasedee Abdullah, Nur Kartinee Bt Kassim, Peter Waziri, and Imaobong Christopher Etti. 2016. "In Silico Discovery of Potential Uridine-Cytidine Kinase 2 Inhibitors from the Rhizome of Alpinia mutica" Molecules 21, no. 4: 417. https://doi.org/10.3390/molecules21040417







