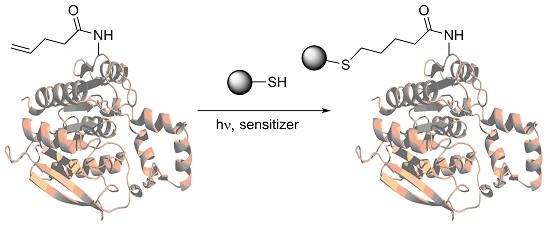
Incorporation of Amino Acids with Long-Chain Terminal Olefins into Proteins
Abstract
:1. Introduction
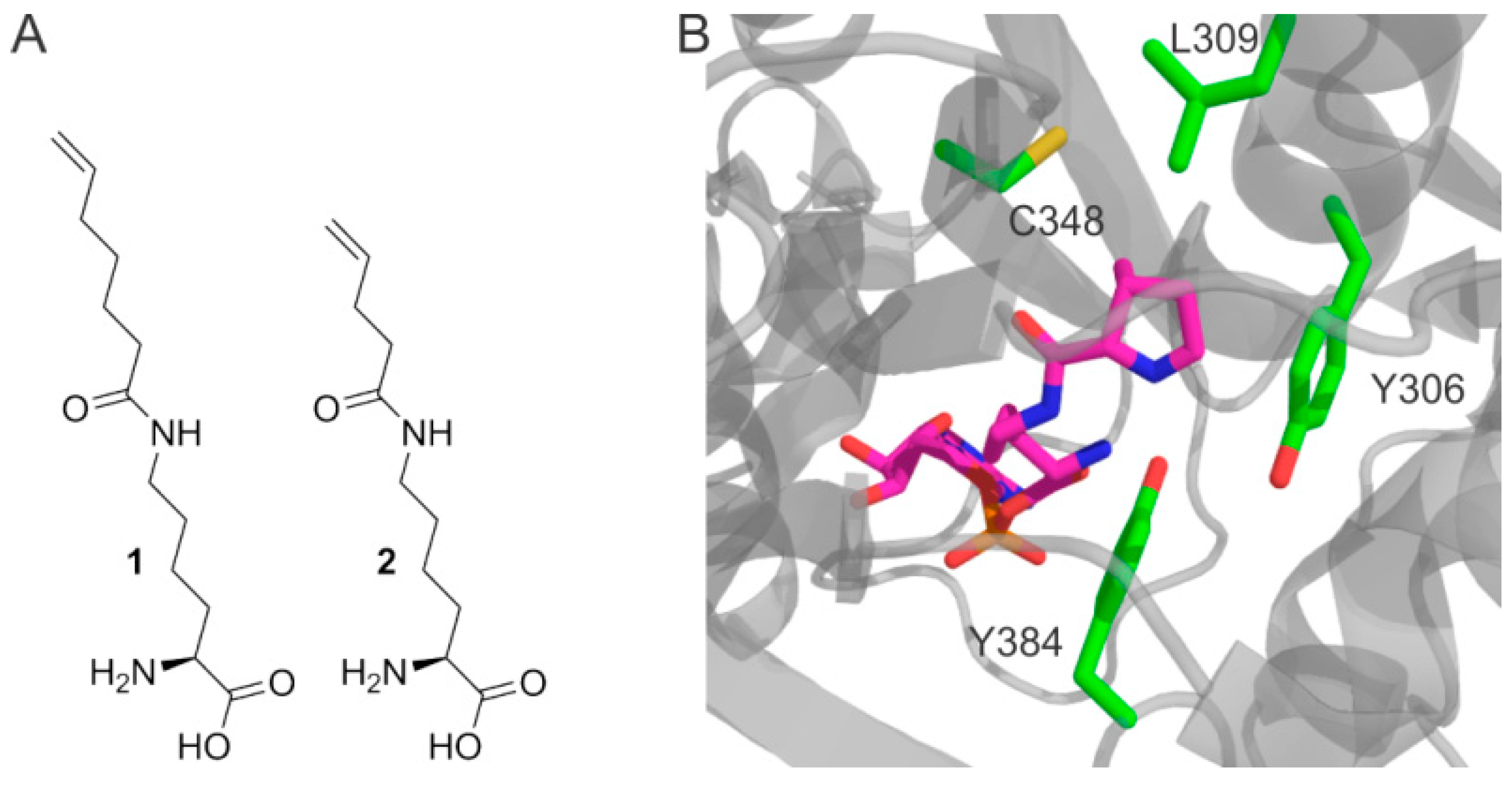
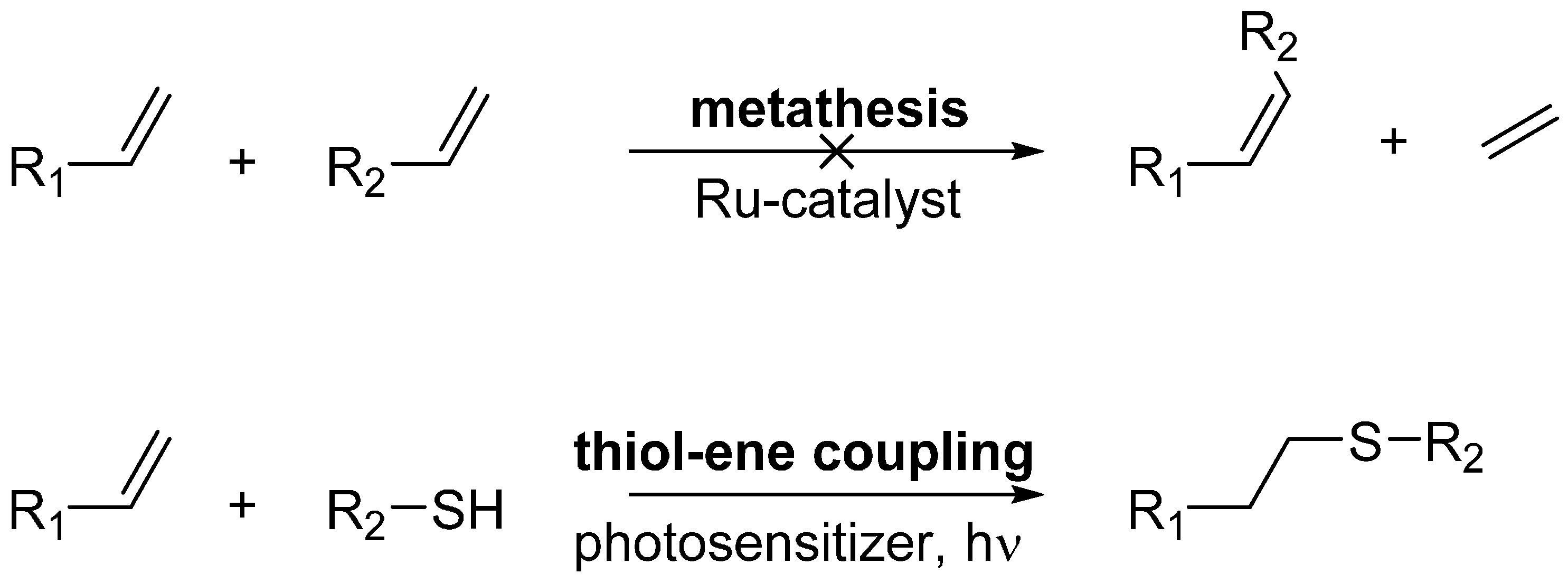
2. Results
3. Discussion
4. Materials and Methods
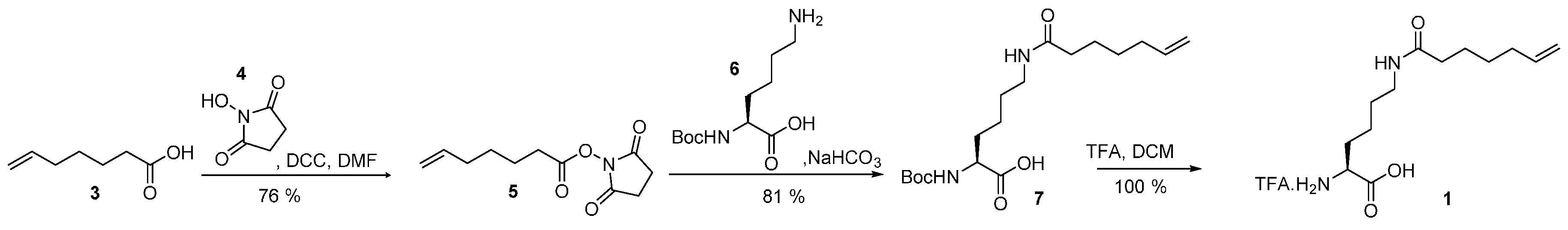
4.1. Synthesis of Hek
4.2. Synthesis of Pek
4.3. Mutagenesis and Expression Tests
4.4. Metathesis Reactions
4.5. Thiol-Ene Coupling
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ncAA: | noncanonical amino acid |
| MmPylRS: | Methanosarcina mazei pyrrolysyl-tRNA synthetase |
| MbPylRS: | Methanosarcina barkeri pyrrolysyl-tRNA synthetase |
| EGFP: | enhanced green fluorescent protein |
| TTL: | Thermoanaerobacter thermohydrosulfuricus lipase |
| Hek: | Nε-heptenoyl lysine |
| Pek: | Nε-pentenoyl lysine |
| HGII: | Hoveyda-Grubbs catalyst, second generation |
References
- Liu, C.C.; Schultz, P.G. Adding New Chemistries to the Genetic Code. Annu. Rev. Biochem. 2010, 79, 413–444. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.R.; Wang, Y.-S.; Wan, W. Synthesis of proteins with defined posttranslational modifications using the genetic noncanonical amino acid incorporation approach. Mol. BioSyst. 2011, 7, 38–47. [Google Scholar] [PubMed]
- Wan, W.; Tharp, J.M.; Liu, W.R. Pyrrolysyl-tRNA synthetase: An ordinary enzyme but an outstanding genetic code expansion tool. Biochim. Biophys. Acta Proteins Proteom. 2014, 1844, 1059–1070. [Google Scholar] [CrossRef] [PubMed]
- Dumas, A.; Lercher, L.; Spicer, C.D.; Davis, B.G. Designing logical codon reassignment—Expanding the chemistry in biology. Chem. Sci. 2015, 6, 50–69. [Google Scholar] [CrossRef]
- Floyd, N.; Vijayakrishnan, B.; Koeppe, J.R.; Davis, B.G. Thiyl Glycosylation of Olefinic Proteins: S-Linked Glycoconjugate Synthesis. Angew. Chem. Int. Ed. 2009, 48, 7798–7802. [Google Scholar]
- Torres-Kolbus, J.; Chou, C.; Liu, J.; Deiters, A. Synthesis of Non-linear Protein Dimers through a Genetically Encoded Thiol-ene Reaction. PLoS ONE 2014, 9, e105467. [Google Scholar]
- Lang, K.; Davis, L.; Wallace, S.; Mahesh, M.; Cox, D.J.; Blackman, M.L.; Fox, J.M.; Chin, J.W. Genetic Encoding of Bicyclononynes and trans-Cyclooctenes for Site-Specific Protein Labeling in Vitro and in Live Mammalian Cells via Rapid Fluorogenic Diels–Alder Reactions. J. Am. Chem. Soc. 2012, 134, 10317–10320. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Wang, Y.; Qu, J.; Lin, Q. Selective Functionalization of a Genetically Encoded Alkene-Containing Protein via “Photoclick Chemistry” in Bacterial Cells. J. Am. Chem. Soc. 2008, 130, 9654–9655. [Google Scholar] [CrossRef] [PubMed]
- Ourailidou, M.E.; van der Meer, J.-Y.; Baas, B.-J.; Jeronimus-Stratingh, M.; Gottumukkala, A.L.; Poelarends, G.J.; Minnaard, A.J.; Dekker, F.J. Aqueous Oxidative Heck Reaction as a Protein-Labeling Strategy. ChemBioChem 2014, 15, 209–212. [Google Scholar] [PubMed]
- Ai, H.-W.; Shen, W.; Brustad, E.; Schultz, P.G. Genetically Encoded Alkenes in Yeast. Angew. Chem. Int. Ed. 2010, 49, 935–937. [Google Scholar]
- Binder, J.B.; Raines, R.T. Olefin metathesis for chemical biology. Curr. Opin. Chem. Biol. 2008, 12, 767–773. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.A.; Chalker, J.M.; Floyd, N.; Bernardes, G.J.L.; Davis, B.G. Allyl Sulfides Are Privileged Substrates in Aqueous Cross-Metathesis: Application to Site-Selective Protein Modification. J. Am. Chem. Soc. 2008, 130, 9642–9643. [Google Scholar] [PubMed]
- Boutureira, O.; Bernardes, G.J.L. Advances in Chemical Protein Modification. Chem. Rev. 2015, 115, 2174–2195. [Google Scholar] [PubMed]
- Van Hest, J.C.; Tirrell, D.A. Efficient introduction of alkene functionality into proteins in vivo. FEBS Lett. 1998, 428, 68–70. [Google Scholar] [PubMed]
- Zhang, Z.; Wang, L.; Brock, A.; Schultz, P.G. The Selective Incorporation of Alkenes into Proteins in Escherichia coli. Angew. Chemie Int. Ed. 2002, 41, 2840–2842. [Google Scholar] [CrossRef]
- Kavran, J.M.; Gundllapalli, S.; O’Donoghue, P.; Englert, M.; Soll, D.; Steitz, T.A. Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation. Proc. Natl. Acad. Sci. USA 2007, 104, 11268–11273. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.-T.; Wang, Y.-S.; Nakamura, A.; Eiler, D.; Kavran, J.M.; Wong, M.; Kiessling, L.L.; Steitz, T.A.; O’Donoghue, P.; Söll, D. Polyspecific pyrrolysyl-tRNA synthetases from directed evolution. Proc. Natl. Acad. Sci. USA 2014, 111, 16724–16729. [Google Scholar] [PubMed]
- Yanagisawa, T.; Ishii, R.; Fukunaga, R.; Kobayashi, T.; Sakamoto, K.; Yokoyama, S. Multistep Engineering of Pyrrolysyl-tRNA Synthetase to Genetically Encode Nε-(o-Azidobenzyloxycarbonyl) lysine for Site-Specific Protein Modification. Chem. Biol. 2008, 15, 1187–1197. [Google Scholar] [PubMed]
- Plass, T.; Milles, S.; Koehler, C.; Schultz, C.; Lemke, E.A. Genetically Encoded Copper-Free Click Chemistry. Angew. Chem. Int. Ed. 2011, 50, 3878–3881. [Google Scholar]
- Schmidt, M.J.; Weber, A.; Pott, M.; Welte, W.; Summerer, D. Structural Basis of Furan-Amino Acid Recognition by a Polyspecific Aminoacyl-tRNA-Synthetase and its Genetic Encoding in Human Cells. ChemBioChem 2014, 15, 1755–1760. [Google Scholar] [PubMed]
- Ogawa, A.; Hayami, M.; Sando, S.; Aoyama, Y. A Concept for Selection of Codon-Suppressor tRNAs Based on Read-Through Ribosome Display in an in Vitro Compartmentalized Cell-Free Translation System. J. Nucleic Acids 2012, 2012, 538129. [Google Scholar] [CrossRef] [PubMed]
- Chalker, J.M.; Lin, Y.A.; Boutureira, O.; Davis, B.G. Enabling olefin metathesis on proteins: chemical methods for installation of S-allyl cysteine. Chem. Commun. 2009, 3714–3716. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.A.; Chalker, J.M.; Davis, B.G. Olefin Cross-Metathesis on Proteins: Investigation of Allylic Chalcogen Effects and Guiding Principles in Metathesis Partner Selection. J. Am. Chem. Soc. 2010, 132, 16805–16811. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.A.; Boutureira, O.; Lercher, L.; Bhushan, B.; Paton, R.S.; Davis, B.G. Rapid Cross-Metathesis for Reversible Protein Modifications via Chemical Access to Se-Allyl-selenocysteine in Proteins. J. Am. Chem. Soc. 2013, 135, 12156–12159. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.A.; Chalker, J.M.; Davis, B.G. Olefin Metathesis for Site-Selective Protein Modification. ChemBioChem 2009, 10, 959–969. [Google Scholar] [PubMed]
- Chalker, J.M. Allyl Sulfides: Reactive Substrates for Olefin Metathesis. Aust. J. Chem. 2015, 68, 1801–1809. [Google Scholar] [CrossRef]
- Köhling, S.; Künze, G.; Lemmnitzer, K.; Bermudez, M.; Wolber, G.; Schiller, J.; Huster, D.; Rademann, J. Chemoenzymatic Synthesis of Nonasulfated Tetrahyaluronan with a Paramagnetic Tag for Studying Its Complex with Interleukin-10. Chem. Eur. J. 2016. [Google Scholar] [CrossRef]
- Hoesl, M.G.; Acevedo-Rocha, C.G.; Nehring, S.; Royter, M.; Wolschner, C.; Wiltschi, B.; Budisa, N.; Antranikian, G. Lipase Congeners Designed by Genetic Code Engineering. ChemCatChem 2011, 3, 213–221. [Google Scholar]
- Budisa, N.; Schulze-Makuch, D. Supercritical Carbon Dioxide and Its Potential as a Life-Sustaining Solvent in a Planetary Environment. Life 2014, 4, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarty, S.P.; Ramapanicker, R.; Mishra, R.; Chandrasekaran, S.; Balaram, H. Development and characterization of lysine based tripeptide analogues as inhibitors of Sir2 activity. Bioorg. Med. Chem. 2009, 17, 8060–8072. [Google Scholar] [CrossRef] [PubMed]
- Al Toma, R.S.; Kuthning, A.; Exner, M.P.; Denisiuk, A.; Ziegler, J.; Budisa, N.; Süssmuth, R.D. Site-Directed and Global Incorporation of Orthogonal and Isostructural Noncanonical Amino Acids into the Ribosomal Lasso Peptide Capistruin. ChemBioChem 2015, 16, 503–509. [Google Scholar] [PubMed]
- Sample Availability: Not available.



© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Exner, M.P.; Köhling, S.; Rivollier, J.; Gosling, S.; Srivastava, P.; Palyancheva, Z.I.; Herdewijn, P.; Heck, M.-P.; Rademann, J.; Budisa, N. Incorporation of Amino Acids with Long-Chain Terminal Olefins into Proteins. Molecules 2016, 21, 287. https://doi.org/10.3390/molecules21030287
Exner MP, Köhling S, Rivollier J, Gosling S, Srivastava P, Palyancheva ZI, Herdewijn P, Heck M-P, Rademann J, Budisa N. Incorporation of Amino Acids with Long-Chain Terminal Olefins into Proteins. Molecules. 2016; 21(3):287. https://doi.org/10.3390/molecules21030287
Chicago/Turabian StyleExner, Matthias P., Sebastian Köhling, Julie Rivollier, Sandrine Gosling, Puneet Srivastava, Zheni I. Palyancheva, Piet Herdewijn, Marie-Pierre Heck, Jörg Rademann, and Nediljko Budisa. 2016. "Incorporation of Amino Acids with Long-Chain Terminal Olefins into Proteins" Molecules 21, no. 3: 287. https://doi.org/10.3390/molecules21030287