Acetaminophen Induced Hepatotoxicity in Wistar Rats—A Proteomic Approach
Abstract
:1. Introduction
2. Results
2.1. Biochemical Analysis
| Enzymes (IU/L) | Acetaminophen (mg/kg) | ||
|---|---|---|---|
| 0 (Control) | 150 | 1500 | |
| Aspartate aminotransferase | 139 ± 4.9 | 115 ± 2.4 | 251 ± 1.2 |
| Alanine aminotransferase | 083 ± 1.5 | 076 ± 1.1 | 130 ± 8.5 |
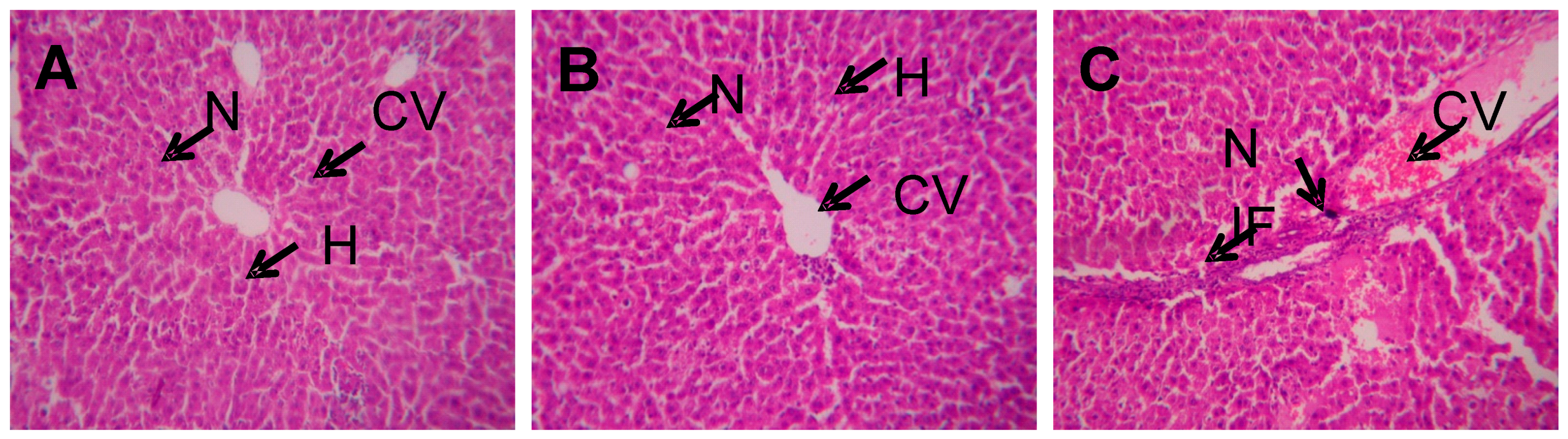
2.2. 2D Gel Analysis

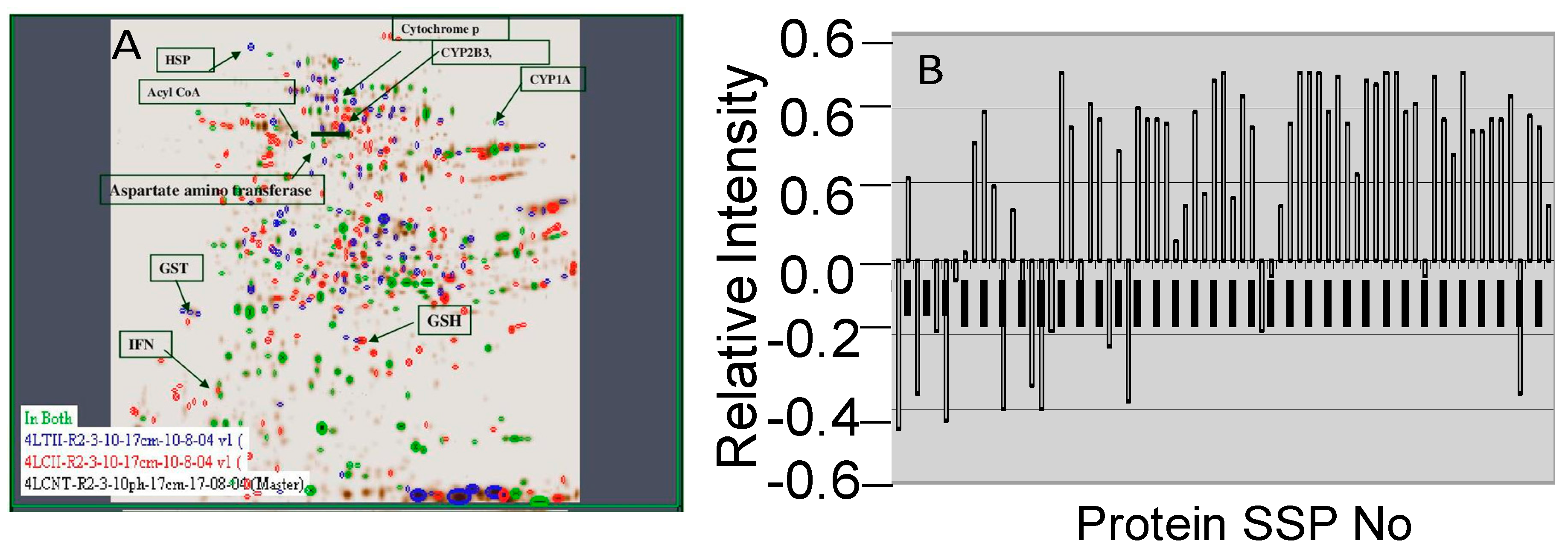
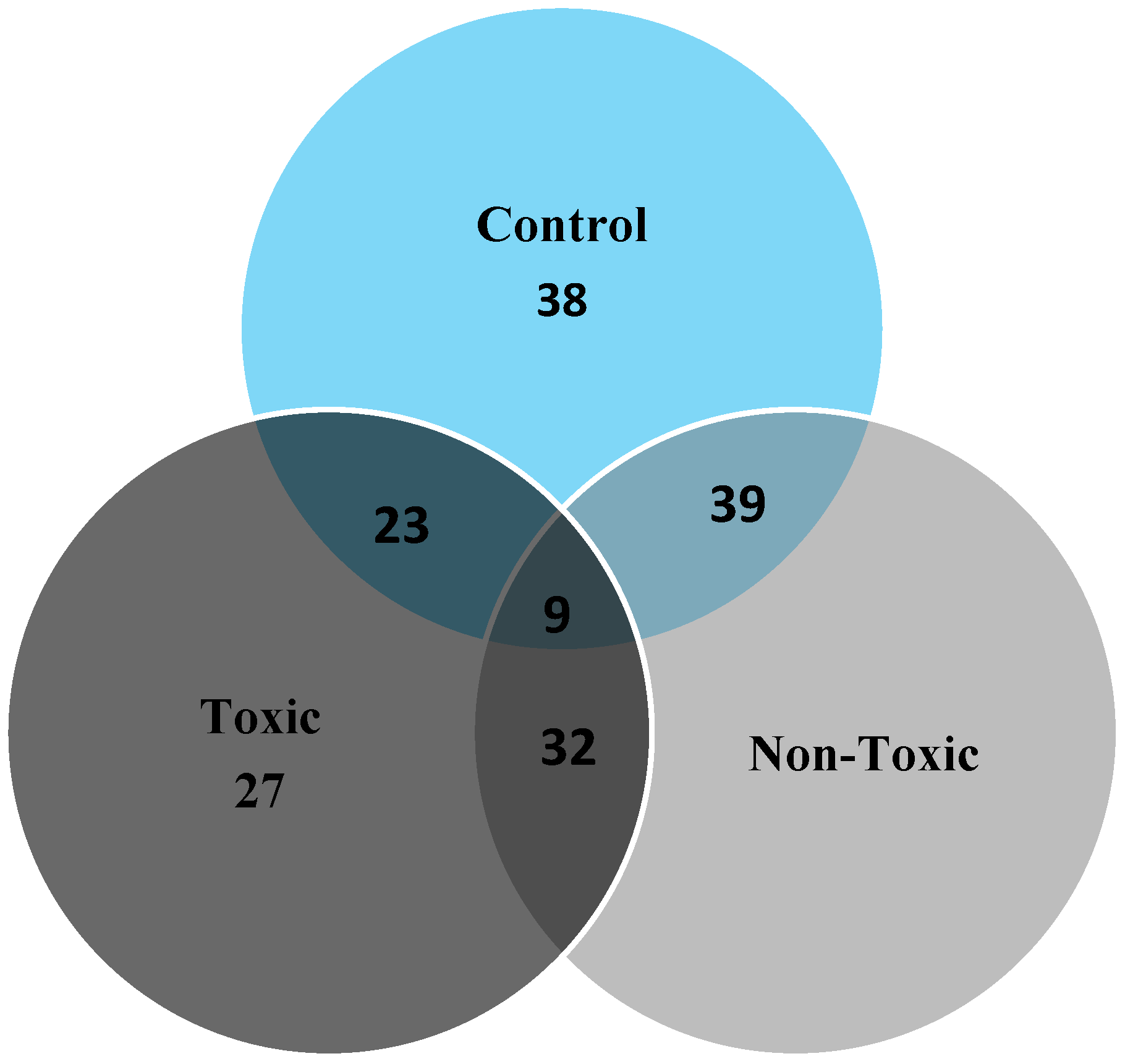
2.3. Image Analysis
2.4. MALDI-TOF Analysis
| S. No | SSP | Protein Name | Sequence Coverage (%) | Estimated M.WT | PI | Swissprot No |
|---|---|---|---|---|---|---|
| 1 | 4514 | Cold-inducible RNA-binding protein | 23 | 38 | 7.0 | P60825 |
| 2 | 3716 | CYP2D3 | 38 | 57 | 6.7 | P12938 |
| 3 | 5910 | Tissue factor pathway inhibitor precursor (TFPI) | 29 | 72 | 7.6 | Q02445 |
| 4 | 2204 | Homeobox protein DRG11 | 21 | 28.6 | 5.9 | Q62798 |
| 5 | 608 | SEC14-like protein 3 | 39 | |||
| 6 | 3 | Low-density lipoprotein receptor-related protein 4 precursor | 42 | 45.1 | 4.8 | Q9Z1J8 |
| 7 | 2308 | Splice isoform 1; Variant Displayed | 27 | 31.6 | 6.1 | Q8VHQ7-00-00-00 |
| 8 | 2206 | Trypsin V-B precursor | 41 | 28 | 6.1 | P32822 |
| 9 | 5 | (NLG1_RAT) Splice isoform | 32 | 13 | Q62765-03-00-00 | |
| 10 | 135 | Somatostatin precursor | 24 | 15.4 | 4.9 | P60042 |
| 11 | 3402 | Guanine nucleotide-binding protein G[I] | 33 | 33 | 6.4 | P54313 |
| 12 | 7602 | Transforming growth factor beta 1 precursor | 38 | 48.2 | 8.4 | P17246 |
| 13 | 4801 | Synaptotagmin X | 47 | 60.7 | 6.8 | O08625 |
| 14 | 7210 | (EPOR_RAT) Splice isoform | 42 | 28.2 | 8.9 | Q07303-01-00-00 |
| 15 | 6412 | Sphingosine 1-phosphate receptor Edg-5 | 39 | 36.6 | 8.2 | P47752 |
| 16 | 3823 | Hydroxymethylglutaryl-CoA synthase, | 35 | 60.6 | 6.8 | P17425 |
| 17 | 5807 | Splice isoform Long; Variant Displayed; | 42 | 61.57 | 7.4 | Q09137-00-00-00 |
| 18 | 8101 | Trefoil factor 2 precursor | 19 | 15.9 | Q09030 | |
| 19 | 2808 | Nonspecific lipid-transfer protein, mitochondrial precursor | 34 | 61 | 6. | P11915 |
| 20 | 6804 | Cyclic-nucleotide-gated olfactory channel | 36 | 61.4 | 7.8 | Q64359 |
| 21 | 3408 | Nuclear transcription factor Y subunit gamma | 29 | 34.4 | 6.5 | Q62725 |
| 22 | 6701 | Neural Wiskott-Aldrich syndrome protein | 35 | 53.2 | 7.8 | O08816 |
| 23 | 8305 | Kinesin heavy chain isoform | 47 | 29 | P56536 | |
| 24 | 2601 | P2X purinoceptor 1 | 16 | 47.9 | 5.8 | P47824 |
| 25 | 5108 | Synaptosomal-associated protein | 41 | 21 | 7.5 | O70377 |
| 26 | 8101 | Phospholipase A2 precursor | 38 | 15.9 | P04055 | |
| 27 | 3409 | 3,2-trans-enoyl-CoA isomerase, | 32 | 35.5 | 6.6 | P23965 |
| 28 | M6 | Cytochrome P450 | 54 | 60.7 | 6.8 | P30839 |
| 29 | 5807 | Growth factor receptor-bound protein 14 | 36 | 61.5 | 7.4 | O88900 |
| 30 | 6710 | Methionine aminopeptidase’s 2 | 49 | 53.2 | 7.8 | P38062 |
| 31 | 1206 | Trypsin I, anionic precursor | 51 | 27.2 | 5.5 | P00762 |
| 32 | 1103 | 60 S Ribosomal protein | 28 | 15.5 | 5.1 | P62907 |
| 33 | 4101 | Peripheral myelin protein 22 | 33 | 11.9 | 6.8 | Q63199 |
| 34 | M1 | GST | 36 | 24.0 | 4.9 | P08011 |
| 35 | 5215 | GSH | 47 | 24 | 7.0 | P19468 |
| S. No | SSP | Protein Name | Sequence Coverage (%) | Score | Estimated M.WT | PI | Swissprot No. |
|---|---|---|---|---|---|---|---|
| 1 | 3716 | CYP2D3 | 25 | 22 | 57 | 6.7 | P12938 |
| 2 | 8305 | Homeobox protein DRG11 | 24 | 19 | 29 | 9.5 | Q62798 |
| 3 | 3705 | 4-Aminobutyrate aminotransferase | 35 | 35 | 56 | 6.4 | P50554 |
| 4 | 3312 | Serine/threonine protein phosphatase | 32 | 34 | 31.5 | 6.7 | P13353 |
| 5 | 6101 | Acyl CoA | 41 | 39 | 18.38 | 7.8 | Q64559 |
| 6 | 5504 | ArgininosPuccinate synthase | 38 | 32 | 43.8 | 7.3 | P09034 |
| 7 | 4801 | Synaptotagmin X | 36 | 38 | 60.7 | 6.8 | O08625 |
| 8 | 8314 | CD82 antigen | 25 | 22 | 30 | O70352 | |
| 9 | 3823 | Hydroxymethylglutaryl-CoA synthase, | 29 | 28 | 60.6 | 6.8 | P17425 |
| 10 | 3306 | F-box/LRR-repeat protein 20 | 40 | 35 | 29.4 | 6.6 | Q9QZH7 |
| 11 | 8301 | Suppressor of cytokine signaling 1 (SOCS-1) | 36 | 32 | 29.2 | 9.1 | Q9QX78 |
| 12 | 1403 | Calcineurin-binding protein Cabin 1 | 34 | 31 | 34.2 | 5.1 | O88480 |
| 13 | 3115 | Hydroxymethylglutaryl-CoA synthase | 33 | 28 | 18.7 | 6.6 | P22791 |
| 14 | 5712 | Lipopolysaccharide-binding protein precursor | 45 | 41 | 56 | 7.5 | Q63313 |
| 15 | 3705 | Cytochrome P450 | 46 | 40 | 56.00 | 6.4 | P20812 |
| 16 | 2908 | Liver carboxylesterase B-1 precursor | 35 | 32 | 66.9 | 6.2 | Q63010 |
| 17 | 5106 | Acetyl-CoA acetyltransferase, mitochondrial precursor | 33 | 34 | 20.4 | 7.5 | P17764 |
| 18 | 3908 | Hyaluronan synthase 2 | 22 | 17 | 66.4 | 6.6 | O35776 |
| 19 | 3522 | Carbonic anhydrase III | 25 | 21 | 38.6 | 6.7 | P14141 |
| 20 | 6019 | Cytochrome c oxidase polypeptide VIa-liver | 29 | 23 | 14.0 | 8.3 | P10818 |
| 21 | 2710 | CYP2B3 | 24 | 20 | 54.8 | 6.3 | P13107 |
| 22 | 4003 | Peripheral myelin protein 22 | 36 | 32 | 11.9 | 6.8 | Q63199 |
| 23 | 8705 | CYP1A2 | 37 | 35 | P04799 | ||
| 24 | M2 | Heat shock protein 70 | 36 | 34 | 70 | 5.8 | Q07439 |
| 25 | M3 | Aspartate aminotransferase | 30 | 33 | 55 | 6.1 | P13221 |
| 26 | M4 | Alanine aminotransferase | 27 | 21 | 54 | P25409 | |
| 27 | M5 | IFN gamma | 23 | 26 | 19 | 4.8 | P01581 |
| 28 | 501 | Mitogen-activated protein kinase | 21 | 20 | 43.4 | 4.9 | Q9WTY9 |
| 29 | 2710 | CYP2B3 | 28 | 24 | 54.8 | 6.3 | P13107 |
| 30 | M6 | Cytochrome P450 | 30 | 24 | 60.7 | 6.8 | P30839 |
| 31 | 8305 | Kinesin heavy chain isoform | 33 | 32 | |||
| 32 | 7802 | (Pyruvate dehydrogenase (Lipoamide))-phosphatase 2 | 38 | 31 | 60 | 8.5 | O88484 |
| 33 | 7602 | Transforming growth factor beta 1 precursor | 36 | 35 | 48.2 | 8.4 | P17246 |
| 34 | 7210 | Calcium-activated potassium channel beta subunit 2 | 41 | 38 | 28 | 8.9 | Q811Q0 |
| 35 | 135 | Hippocalcin-like protein | 52 | 47 | 15 | 4.9 | P62749 |
| 36 | 4002 | Cadherin-14 (Fragment) (Rat) | 17 | 21 | 11.0 | 6.8 | Q9Z2V8 |
| 37 | 3 | Single-stranded DNA-binding protein | 20 | 25 | 14 | P28042 | |
| 38 | 850 | Splice isoform Displayed | 24 | 17 | 43.4 | 4.9 | Q9R0E0 |
| 39 | 3512 | Wnt-5a protein precursor | 35 | 34 | 41.7 | 6.5 | Q9QXQ7 |
| 40 | 1204 | 60S ribosomal protein L7 | 21 | 20 | 23 | 5.3 | P37805 |
| 41 | 3823 | Phosphoglucomutase | 34 | 32 | 60.6 | 6.8 | P38652 |
| 42 | 8705 | Glutamate decarboxylase | 36 | 34 | 60.5 | 9.2 | P18088 |

| S. No | Protein Identified | Swiss Port NO | M.WT (Kda) |
|---|---|---|---|
| 1 | Cytochrome P450 | P20812 | 56.00 |
| 2 | Heat shock protein70 | Q07439 | 70 |
| 3 | IFN-γ | P01581 | 19 |
| 4 | Cyp2B3 | P13107 | 54.8 |
| 5 | Cyp2D3 | P12938 | 57 |
| 6 | AST | P13221 | 55 |
| 7 | ALT | P25409 | 54 |
| 8 | Suppressor of cytokine signaling 1 | Q9QX78 | 29.2 |
| 9 | Liver carboxylesterase B-1 precursor | Q63010 | 66.9 |
| 10 | Acetyl-CoA acetyltransferase, mitochondrial precursor | P17764 | 20.4 |
| 11 | Cytochrome c oxidase polypeptide VIa-liver | P10818 | 14.0 |
| 12 | Calcineurin-binding protein Cabin 1 | O88480 | 34.2 |
| 13 | Hydroxymethylglutaryl-CoA synthase | 18.7 | P22791 |
3. Discussion
4. Experimental Section
4.1. Animal Treatment
4.2. Biochemical Analysis
4.3. Histopathology Analysis
4.4. Protein Sample Preparation
4.5. DE and Image Analysis
4.6. In-gel Tryptic Digestion and Mass Spectrometry (MALDI-TOF) Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Boutis, K.; Shannon, M. Nephrotoxicity after acute severe acetaminophen poisoning in adolescents. Clin. Toxicol. 2001, 39, 441–445. [Google Scholar] [CrossRef]
- Davidson, D.G.; Eastham, W.N. Acute liver necrosis following overdose of paracetamol. Br. Med. J. 1996, 5512, 497–499. [Google Scholar] [CrossRef]
- Kleinman, J.G.; Breitenfield, R.V.; Roth, D.A. Acute renal failure associated with acetaminophen ingestion:Report of a case and review of the literature. Clin. Nephrol. 1980, 14, 201–205. [Google Scholar] [PubMed]
- Mudge, G.H.; Gemborys, M.W.; Duggin, G.G. Covalent binding of metabolites of acetaminophen to kidney protein and depletion of renal glutathione. J. Pharmacol. Exp. Ther. 1978, 206, 218–226. [Google Scholar]
- Vidal, B.C.; Bonventre, J.V.; I-Hong Hsu, S. Towards the application of proteomics in renal disease in renal disease. Clin. Sci. 2005, 109, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Pixton, K.L.; Deeks, E.D.; Flesch, F.M. Sperm proteome mapping of a patient who experienced failed fertilization at IVF reveals altered expression of at least 20 proteins compared with fertile donors: case report. Hum. Reprod. 2004, 19, 1438–1447. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.A.; Witherdin, R.; Hetherington, L. Identification of posttranslational modifications that occur during sperm maturation using difference in two-dimensional gel electrophoresis. Proteomics 2005, 5, 1003–1012. [Google Scholar] [CrossRef] [PubMed]
- Naaby-Hansen, S.; Mandal, A.; Wolkowicz, M.J.; Sen, B.; Westbrook, V.A.; Shetty, J.; Coonrod, S.A.; Klotz, K.L.; Kim, Y.H.; Bush, L.A.; et al. CABYR, a novel calcium-binding tyrosine phosphorylation-regulated fibrous sheath protein involved in capacitation. Dev. Biol. 2002, 242, 236–254. [Google Scholar] [CrossRef] [PubMed]
- Turner, R.M.; Musse, M.P.; Mandal, A. Molecular genetic analysis of two human sperm fibrous sheath proteins, AKAP4 and AKAP3, in men with dysplasia of the fibrous sheath. J. Androl. 2001, 22, 302–315. [Google Scholar] [PubMed]
- Bohring, C.; Krause, W. Immune infertility: towards a better understanding of sperm [auto]-immunity. The value of proteomic analysis. Hum. Reprod. 2003, 18, 915–924. [Google Scholar] [CrossRef] [PubMed]
- Nandhini, V.S.; Viji Stella Bai, G. In-vitro phytopharmacological effect and cardio protective activity of Rauvolfia tetraphylla L. South Ind. J. Biol. Sci. 2015, 1, 97–102. [Google Scholar]
- Kleiner, D.E.; Brunt, E.M.; van Natta, M.; Behling, C.; Contos, M.J.; Cummings, O.W.; Ferrell, L.D.; Liu, Y.C.; Torbenson, M.S.; Unalp-Arida, A.; et al. Non alcoholic steato hepatitis clinical research network, design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 2005, 41, 1313–1321. [Google Scholar] [PubMed]
- Dorne, J.L.; Skinner, L.; Frampton, G.K.; Spurgeon, D.J.; Ragas, A.M. Human and environmental risk assessment of pharmaceuticals: differences, similarities, lessons from toxicology. Anal. Bioanal. Chem. 2007, 387, 1259–1268. [Google Scholar] [CrossRef] [PubMed]
- Burke, W.; Psaty, B.M. Personalized medicine in the era of genomics. JAMA 2007, 298, 1682–1684. [Google Scholar] [CrossRef] [PubMed]
- Rajesh, M.G.; Latha, M.S. Protective effect of Glycyrrhiza glabra Linn. on carbon tetrachloride-induced peroxidative damage. Ind. J. Pharmacol. 2004, 36, 284–286. [Google Scholar]
- Martin, J.; Smilkstein, M.D.; Gary, L.; Knapp, M.S.; Kenneth, W.; Kulig, M.D.; Barry, H.; Rumack, M.D. Efficacy of Oral N-Acetylcysteine in the Treatment of Acetaminophen Overdose. N. Engl. J. Med. 1988, 319, 1557–1562. [Google Scholar]
- Corcoran, G.B.; Mitchell, J.R.; Vaishnav, Y.N.; Horning, E.C. Evidence that acetaminophen and N-hydroxyacetaminophen form a common arylating intermediate, N-acetyl-p-benzo-quinoneimine. Mol. Pharmacol. 1980, 18, 536–542. [Google Scholar] [PubMed]
- Dahlin, D.C.; Miwa, G.T.; Lu, A.Y.; Nelson, S.D. N-acetyl-pbenzoquinone imine: a cytochrome P-450-mediated oxidation product of acetaminophen. Proc. Natl. Acad. Sci. USA 1984, 81, 1327–1331. [Google Scholar] [CrossRef] [PubMed]
- Manautou, J.E.; Emeigh Hart, S.G.; Khairallah, E.A.; Cohen, S.D. Protection against acetaminophen hepatotoxicity by a single dose of clofibrate: effects on selective protein arylation and glutathione depletion. Fundam. Appl. Toxicol. 1996, 29, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Rashed, M.S.; Myers, T.G.; Nelson, S.D. Hepatic protein arylation, glutathione depletion, and metabolite profiles of acetaminophen and a non-hepatotoxic regioisomer, 3’-hydroxyacetanilide, in the mouse. Drug Metab. Dispos. 1990, 18, 765–770. [Google Scholar] [PubMed]
- Bartolone, J.B.; Birge, R.B.; Bulera, S.J.; Bruno, M.K.; Nishanian, E.V.; Cohen, S.D.; Khairallah, E.A. Purification, antibody production, and partial amino acid sequence of the 58-kDa acetaminophen-binding liver proteins. Toxicol. Appl. Pharmacol. 1992, 113, 19–29. [Google Scholar] [CrossRef]
- Bulera, S.J.; Birge, R.B.; Cohen, S.D.; Khairallah, E.A. Identification of the mouse liver 44-kDa acetaminophen-binding protein as a subunit of glutamine synthetase. Toxicol. Appl. Pharmacol. 1995, 134, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds acetaminophen are available from the authors. Also it can be purchased from Sigma-Aldrich.
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ilavenil, S.; Al-Dhabi, N.A.; Srigopalram, S.; Ock Kim, Y.; Agastian, P.; Baru, R.; Choi, K.C.; Valan Arasu, M. Acetaminophen Induced Hepatotoxicity in Wistar Rats—A Proteomic Approach. Molecules 2016, 21, 161. https://doi.org/10.3390/molecules21020161
Ilavenil S, Al-Dhabi NA, Srigopalram S, Ock Kim Y, Agastian P, Baru R, Choi KC, Valan Arasu M. Acetaminophen Induced Hepatotoxicity in Wistar Rats—A Proteomic Approach. Molecules. 2016; 21(2):161. https://doi.org/10.3390/molecules21020161
Chicago/Turabian StyleIlavenil, Soundharrajan, Naif Abdullah Al-Dhabi, Srisesharam Srigopalram, Young Ock Kim, Paul Agastian, Rajasekhar Baru, Ki Choon Choi, and Mariadhas Valan Arasu. 2016. "Acetaminophen Induced Hepatotoxicity in Wistar Rats—A Proteomic Approach" Molecules 21, no. 2: 161. https://doi.org/10.3390/molecules21020161





