cDNA Isolation and Functional Characterization of UDP-d-glucuronic Acid 4-Epimerase Family from Ornithogalum caudatum
Abstract
:1. Introduction
2. Results and Discussion
2.1. Unigenes Retrieval from O. caudatum RNA-Seq Data
2.2. cDNA Isolation of OcUGlcAE Family
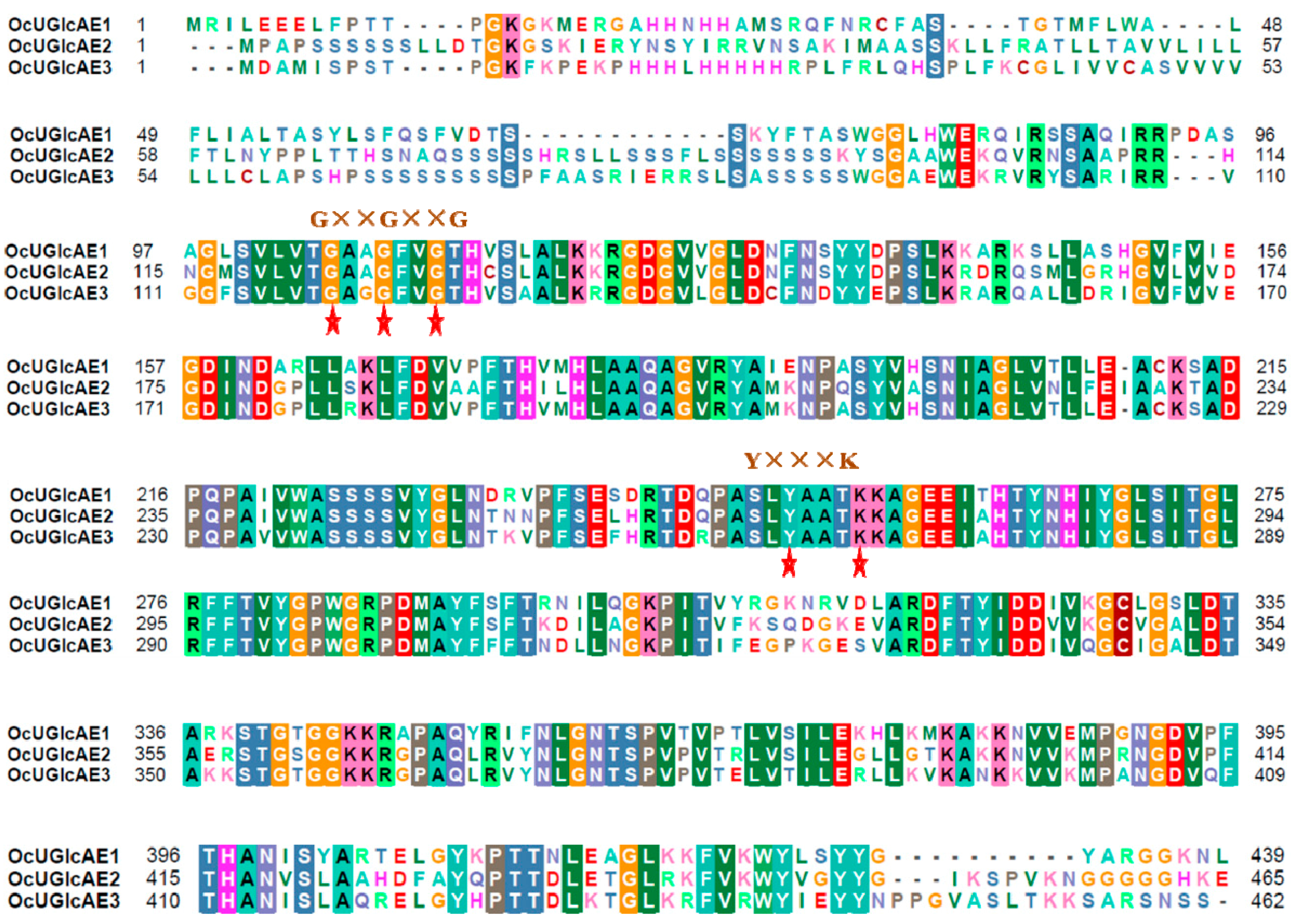
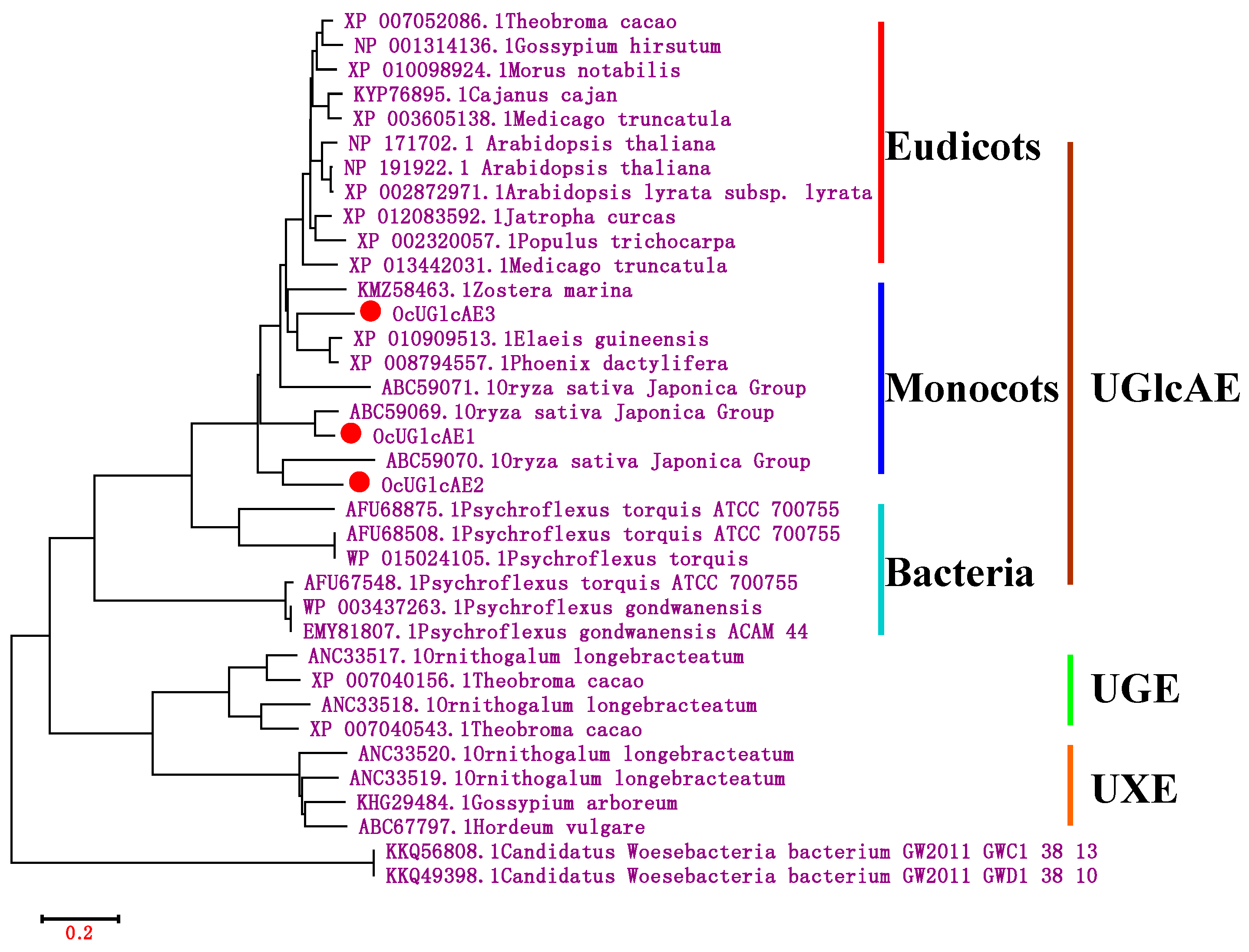
2.3. Bioinformatics Characterization of OcUGlcAE Family
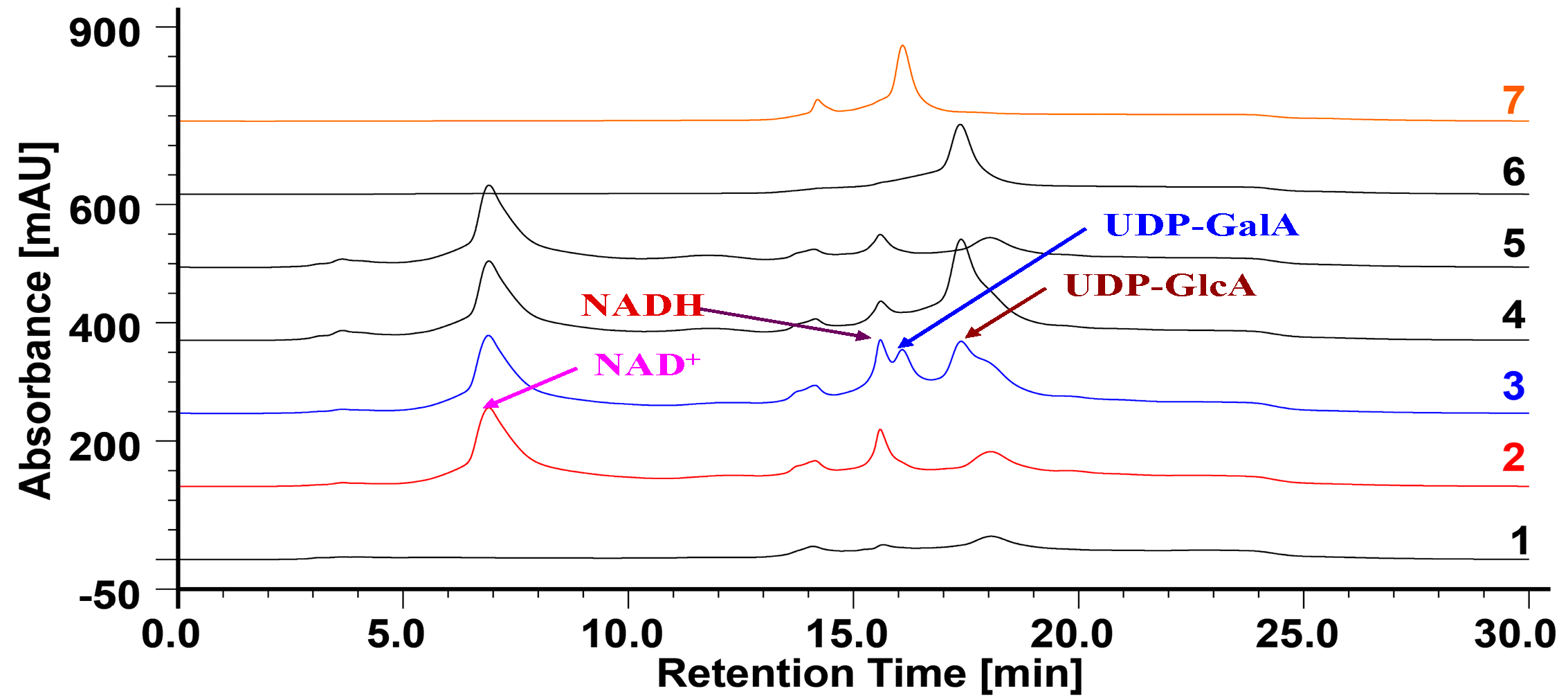
2.4. Bacterial Expression of OcUGlcAEs
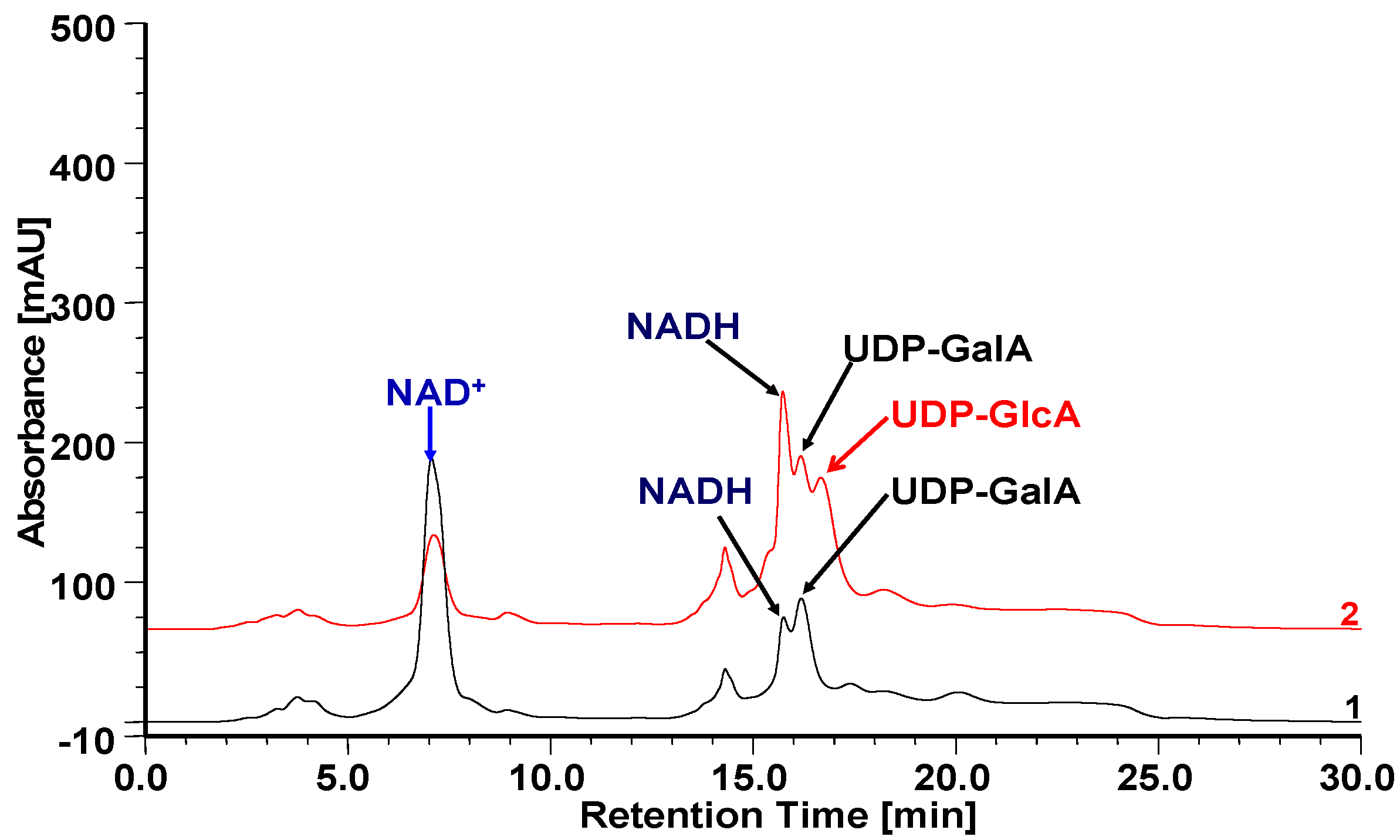
2.5. Heterologous Expression of OcUGlcAEs in P. pastoris
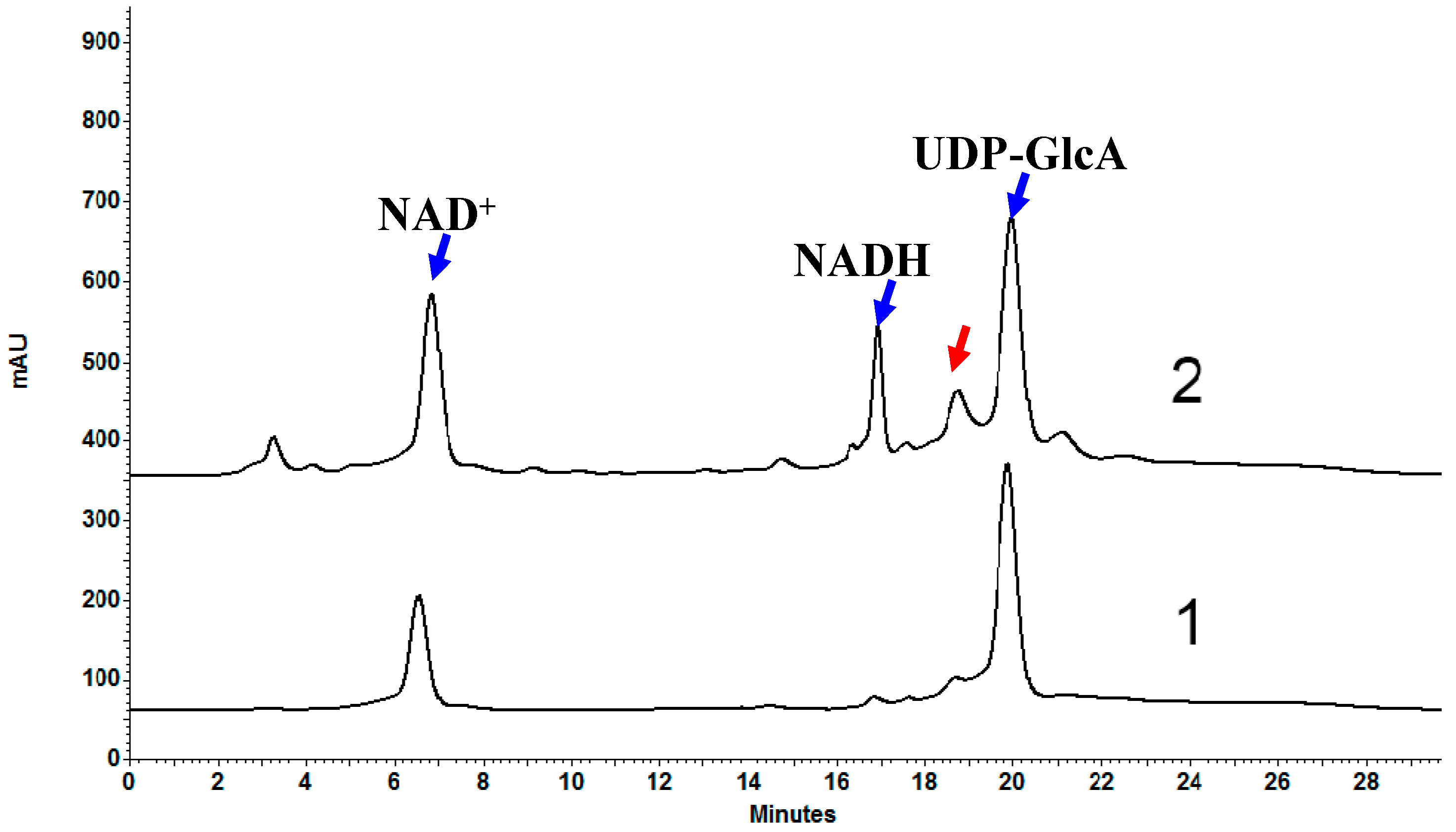
2.6. Biochemical Properties of OcUGlcAE3
2.7. Expression Analyses of OcUGlcAE in O. caudatum
2.8. Discussion
3. Materials and Methods
3.1. Plasmids, Strains and Compounds
3.2. Bacterial and Yeast Culture Media
3.3. Plant Material
3.4. Unigenes Retrieval from O. caudatum RNA-Seq Data
3.5. cDNA Isolation and Bioinformatics Analyses of OcUGlcAEs
3.6. Bacterial Expression of OcUGlcAEs
3.7. Yeast Expression and Functional Characterization of OcUGlcAE1
3.8. Enzymatic Characterization of OcUGlcAE3
3.9. Expression Analyses of OcUGlcAEs in O. caudatum
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| BMGY | buffered minimal glycerol-complex medium |
| GalA | d-galacturonic acid |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| HPLC | High performance liquid chromatography |
| MD | Minimal dextrose |
| ORF | open reading frame |
| PBS | Phosphate buffered saline |
| RT-qPCR | real-time quantitative PCR |
| SDR | short-chain dehydrogenase/reductase |
| trun-OcUGlcAEs | Truncated OcUGlcAE proteins |
| UDP-Ara | UDP-l-arabinose |
| UDP-Gal | UDP-d-galactose |
| UDP-GalA | UDP-d-galacturonic acid |
| UDP-Glc | UDP-d-glucose |
| UDP-GlcA | UDP-d-glucuronic acid |
| UDP-Xyl | UDP-d-xylose |
| UGlcAE | UDP-d-glucuronic acid 4-epimerase |
| UGE | UDP-d-glucose 4-epimerase |
| UTR | untranslated region |
| UXE | UDP-d-xylose 4-epimerase |
| YPD | yeast extract–peptone–dextrose medium |
References
- Yin, Y.; Huang, J.; Gu, X.; Bar-Peled, M.; Xu, Y. Evolution of plant nucleotide-sugar interconversion enzymes. PLoS ONE 2011, 6, e27995. [Google Scholar] [CrossRef] [PubMed]
- Bar-Peled, M.; O’Neill, M.A. Plant nucleotide sugar formation, interconversion, and salvage by sugar recycling. Annu. Rev. Plant Biol. 2011, 62, 127–155. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Bar-Peled, M. The biosynthesis of UDP-galacturonic acid in plants. Functional cloning and characterization of Arabidopsis UDP-d-glucuronic acid 4-epimerase. Plant Physiol. 2004, 136, 4256–4264. [Google Scholar] [CrossRef] [PubMed]
- Broach, B.; Gu, X.; Bar-Peled, M. Biosynthesis of UDP-glucuronic acid and UDP-galacturonic acid in Bacillus cereus subsp. cytotoxis NVH 391-98. FEBS J. 2012, 279, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Wages, C.J.; Davis, K.E.; Guyett, P.J.; Bar-Peled, M. Enzymatic characterization and comparison of various poaceae UDP-GlcA 4-epimerase isoforms. J. Biochem. 2009, 146, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yu, D.Q. Two new constituents from Erigeron breviscapus. J. Asian. Nat. Prod. Res. 2013, 15, 969–973. [Google Scholar] [CrossRef] [PubMed]
- Eshbakova, K.A.; Toshmatov, Z.O.; Yili, A.; Aisa, H.A.; Abdullaev, N.D. Flavonoid galacturonides and glucuronide from the aerial part of Scutellaria schachristanica. Chem. Nat. Compd. 2013, 49, 103–105. [Google Scholar] [CrossRef]
- El-Kassem, L.T.A.; Mohammed, R.S.; El Souda, S.S.; El-Anssary, A.A.; Hawas, U.W.; Mohmoud, K.; Farrag, A.R. Digalacturonide flavones from Egyptian Lantana camara flowers with in vitro antioxidant and in vivo hepatoprotective activities. Z. Naturforsch. C 2012, 67, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Salama, M.M.; Ezzat, S.M.; Sleem, A.A. A new hepatoprotective flavone glycoside from the flowers of Onopordum alexandrinum growing in Egypt. Z. Naturforsch. C 2011, 66, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Frirdich, E.; Whitfield, C. Characterization of Gla(KP), a UDP-galacturonic acid C4-epimerase from Klebsiella pneumoniae with extended substrate specificity. J. Bacteriol. 2005, 187, 4104–4115. [Google Scholar] [CrossRef] [PubMed]
- Keating, D.H.; Willits, M.G.; Long, S.R. A Sinorhizobium meliloti lipopolysaccharide mutant altered in cell surface sulfation. J. Bacteriol. 2002, 184, 6681–6689. [Google Scholar] [CrossRef] [PubMed]
- Paton, A.W.; Paton, J.C. Molecular characterization of the locus encoding biosynthesis of the lipopolysaccharide O antigen of Escherichia coli serotype O113. Infect. Immun. 1999, 67, 5930–5937. [Google Scholar] [PubMed]
- Muñoz, R.; López, R.; de Frutos, M.; García, E. First molecular characterization of a uridine diphosphate galacturonate 4-epimerase: An enzyme required for capsular biosynthesis in Streptococcus pneumoniae type 1. Mol. Microbiol. 1999, 31, 703–713. [Google Scholar] [CrossRef] [PubMed]
- Usadel, B.; Schluter, U.; Molhoj, M.; Gipmans, M.; Verma, R.; Kossmann, J.; Reiter, W.D.; Pauly, M. Identification and characterization of a UDP-d-glucuronate 4-epimerase in Arabidopsis. FEBS Lett. 2004, 569, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Molhoj, M.; Verma, R.; Reiter, W.D. The biosynthesis of d-galacturonate in plants. functional cloning and characterization of a membrane-anchored UDP-d-glucuronate 4-epimerase from Arabidopsis. Plant Physiol. 2004, 135, 1221–1230. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Meng, F.; Liu, Z.; Chen, R.; Zhang, M. Antitumor activities of different fractions of polysaccharide purified from Ornithogalum caudatum Ait. Carbohyd. Polym. 2010, 80, 845–851. [Google Scholar] [CrossRef]
- Yin, S.; Kong, J.-Q. Transcriptome-guided discovery and functional characterization of two UDP-sugar 4-epimerase families involved in the biosynthesis of anti-tumor polysaccharides in Ornithogalum caudatum. RSC Adv. 2016, 6, 37370–37384. [Google Scholar] [CrossRef]
- Li, L.-N.; Kong, J.Q. Transcriptome-wide identification of sucrose synthase genes in Ornithogalum caudatum. RSC Adv. 2016, 6, 18778–18792. [Google Scholar] [CrossRef]
- Guo, L.; Chen, X.; Li, L.-N.; Tang, W.; Pan, Y.-T.; Kong, J.-Q. Transcriptome-enabled discovery and functional characterization of enzymes related to (2S)-pinocembrin biosynthesis from Ornithogalum caudatum and their application for metabolic engineering. Microb. Cell Fact. 2016, 15, 27. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, L.-N.; Pan, Y.-T.; Kong, J.-Q. cDNA isolation and functional characterization of squalene synthase gene from Ornithogalum caudatum. Protein Expr. Purif. 2017, 130, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Lilie, H.; Schwarz, E.; Rudolph, R. Advances in refolding of proteins produced in E. coli. Curr. Opin. Biotechnol. 1998, 9, 497–501. [Google Scholar] [CrossRef]
- Clark, E.D.B. Refolding of recombinant proteins. Curr. Opin. Biotechnol. 1998, 9, 157–163. [Google Scholar] [CrossRef]
- Yin, S.; Kong, J.-Q. Transcriptome-guided gene isolation and functional characterization of UDP-xylose synthase and UDP-d-apiose/UDP-d-xylose synthase families from Ornithogalum caudatum Ait. Plant Cell Rep. 2016, 35, 2403–2421. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-B.; Chen, X.; Wang, W.; Cheng, K.-D.; Kong, J.-Q. Transcriptome-wide identification and characterization of Ornithogalum saundersiae phenylalanine ammonia lyase gene family. RSC Adv. 2014, 4, 27159–27175. [Google Scholar] [CrossRef]
- Sample Availability: OcUGlcAE3 gene and corresponding plasmids are available from the authors.

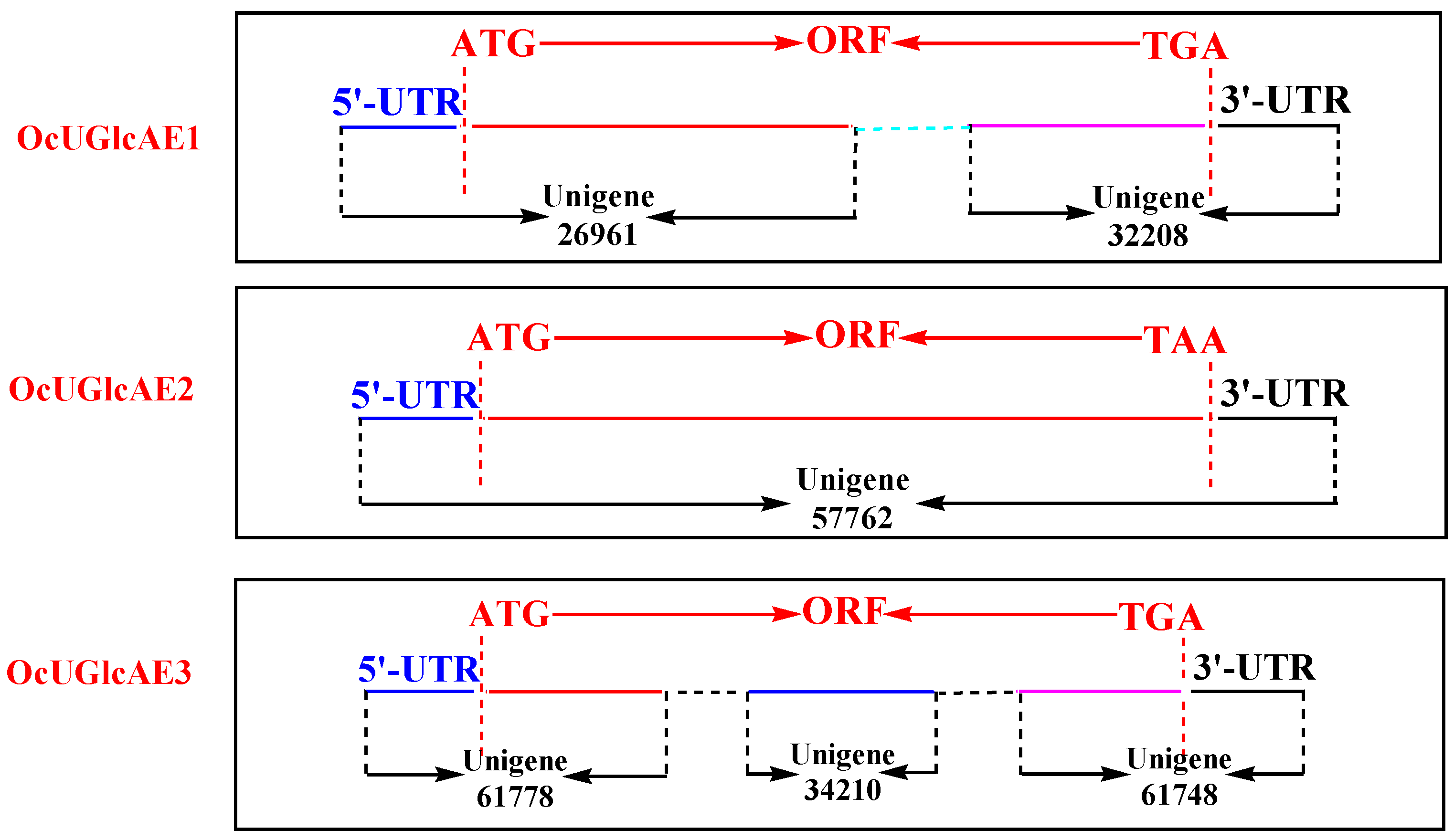



| Unigene ID | Length (bp) | Sequence Structure (bp) | ||
|---|---|---|---|---|
| 5’-UTR | ORF | 3’-UTR | ||
| 26961 | 1011 | 206 | 805 * | N |
| 32208 | 634 | N | 459 * | 175 |
| 57762 | 2247 | 270 | 1398 | 579 |
| 34210 | 383 | N | 383 * | N |
| 61748 | 664 | N | 354 * | 310 |
| 61778 | 478 | 148 | 330 * | N |
| Nucleotide | Protein | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Accession Number | Corresponding Unigene (s) | Length (bp) | ORF (bp) | a.a. | kDa | pI | Identity (%) | |
| OcUGlcAE2 | OcUGlcAE3 | ||||||||
| OcUGlcAE1 | KX429689 | Unigenes 26961 and 32208 | 1701 | 1320 | 439 | 48.462 | 9.66 | 74 | 76 |
| OcUGlcAE2 | KX429690 | Unigene 57762 | 2247 | 1398 | 465 | 50.057 | 9.76 | 100 | 77 |
| OcUGlcAE3 | KX429691 | Unigenes 61778, 61748 and 34210 | 1847 | 1389 | 463 | 50.579 | 9.77 | 77 | 100 |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, S.; Sun, Y.-J.; Liu, M.; Li, L.-N.; Kong, J.-Q. cDNA Isolation and Functional Characterization of UDP-d-glucuronic Acid 4-Epimerase Family from Ornithogalum caudatum. Molecules 2016, 21, 1505. https://doi.org/10.3390/molecules21111505
Yin S, Sun Y-J, Liu M, Li L-N, Kong J-Q. cDNA Isolation and Functional Characterization of UDP-d-glucuronic Acid 4-Epimerase Family from Ornithogalum caudatum. Molecules. 2016; 21(11):1505. https://doi.org/10.3390/molecules21111505
Chicago/Turabian StyleYin, Sen, Yu-Jia Sun, Ming Liu, Li-Na Li, and Jian-Qiang Kong. 2016. "cDNA Isolation and Functional Characterization of UDP-d-glucuronic Acid 4-Epimerase Family from Ornithogalum caudatum" Molecules 21, no. 11: 1505. https://doi.org/10.3390/molecules21111505







