Group I Intron Internal Guide Sequence Binding Strength as a Component of Ribozyme Network Formation
Abstract
:1. Introduction
2. Results
2.1. Experimental Design
2.1.1. Fluorescence Anisotropy
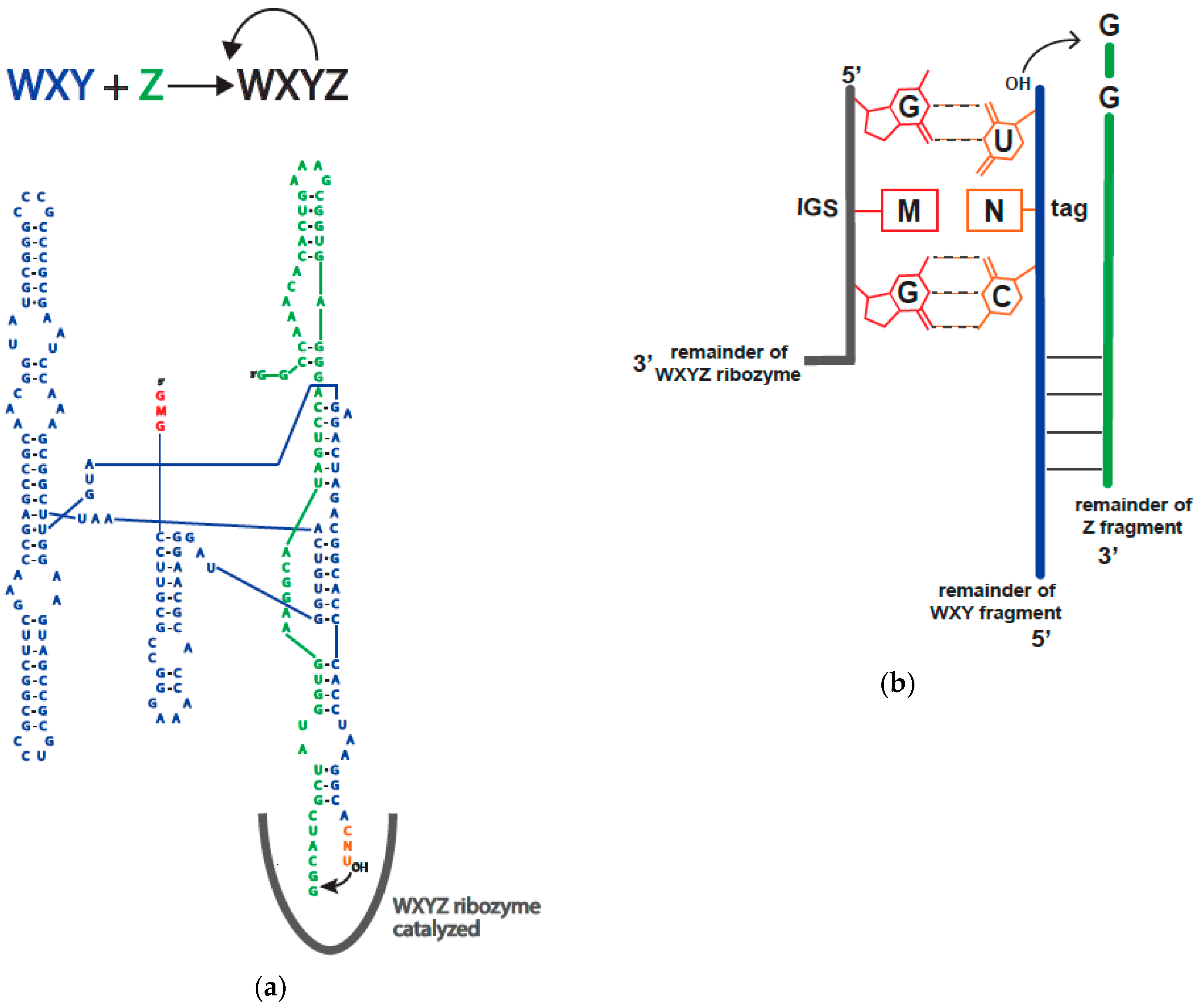
2.1.2. Model System
2.2. Catalytic Abilities of the 16 Genotypes
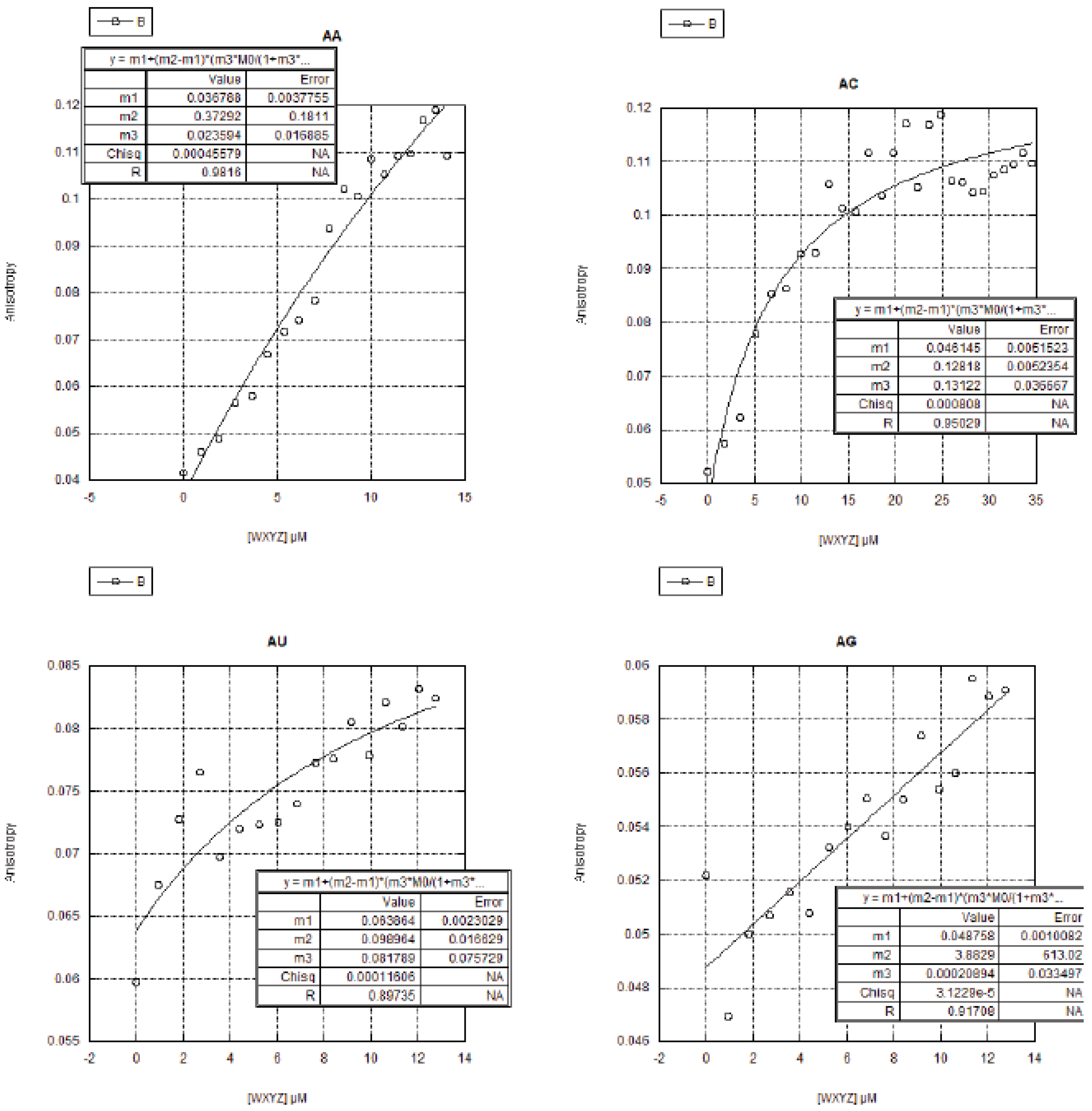
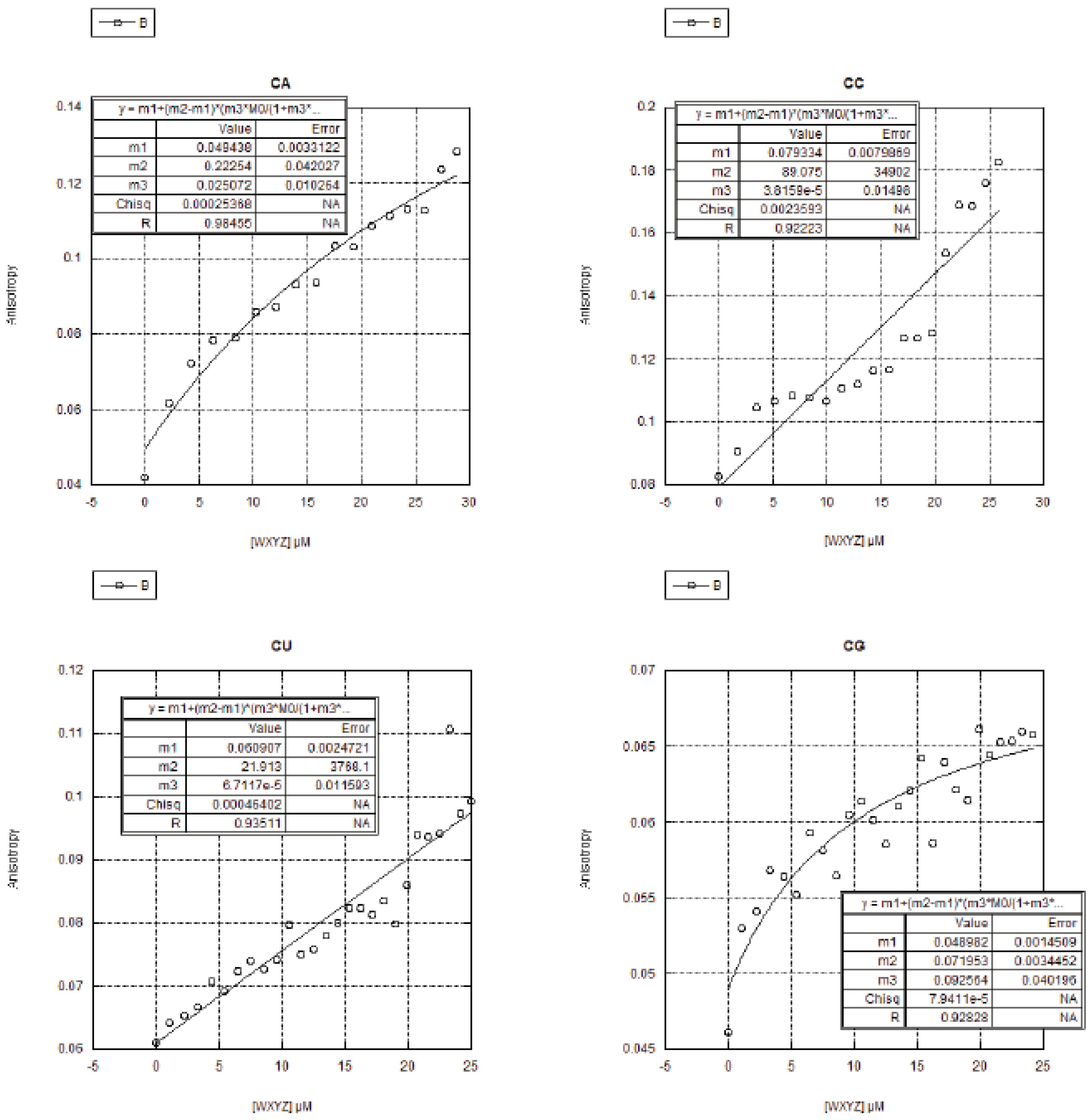
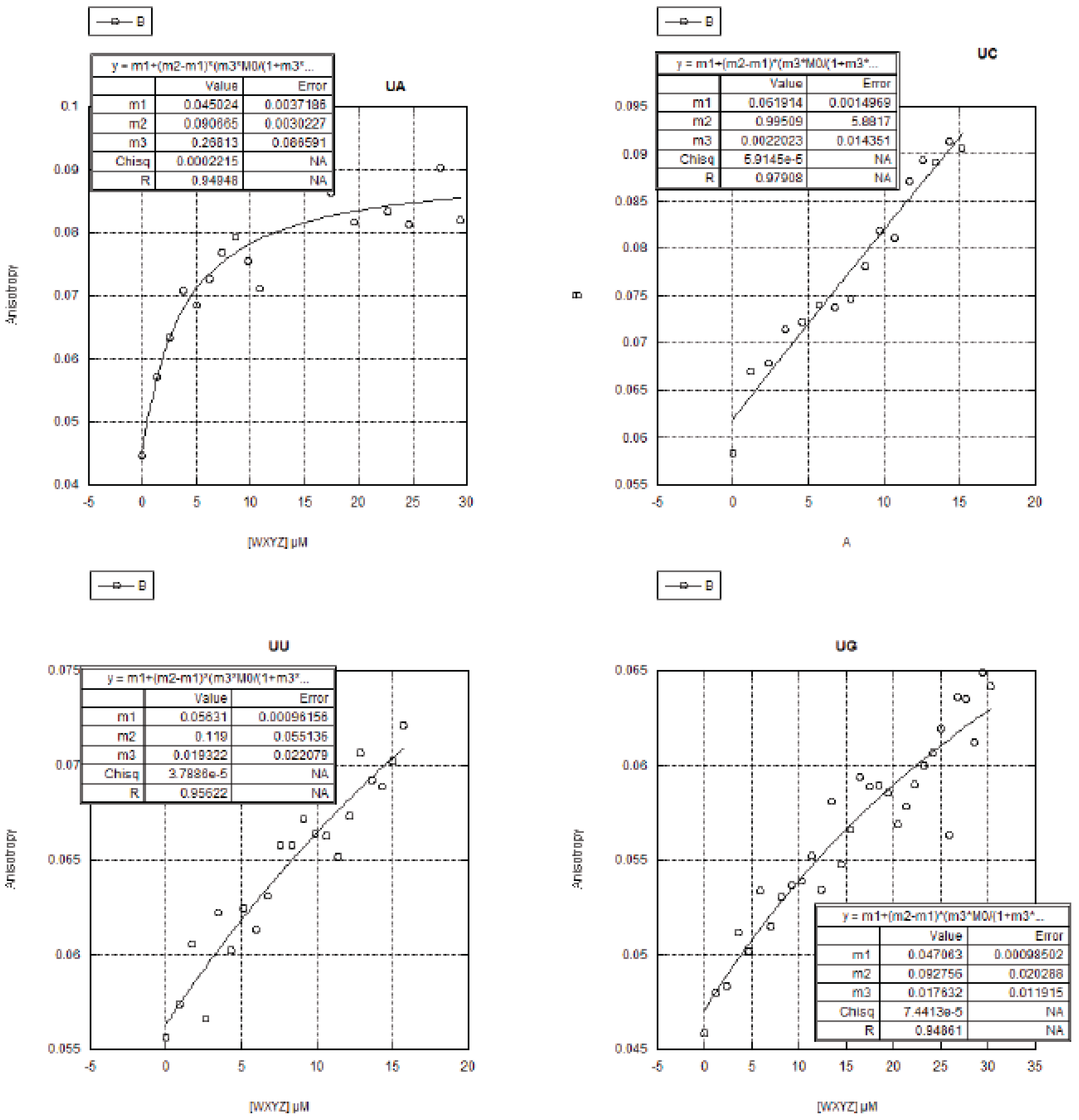
2.3. FA Measurements of the 16 Genotypes
2.3.1. Curve Fitting
2.3.2. Trends
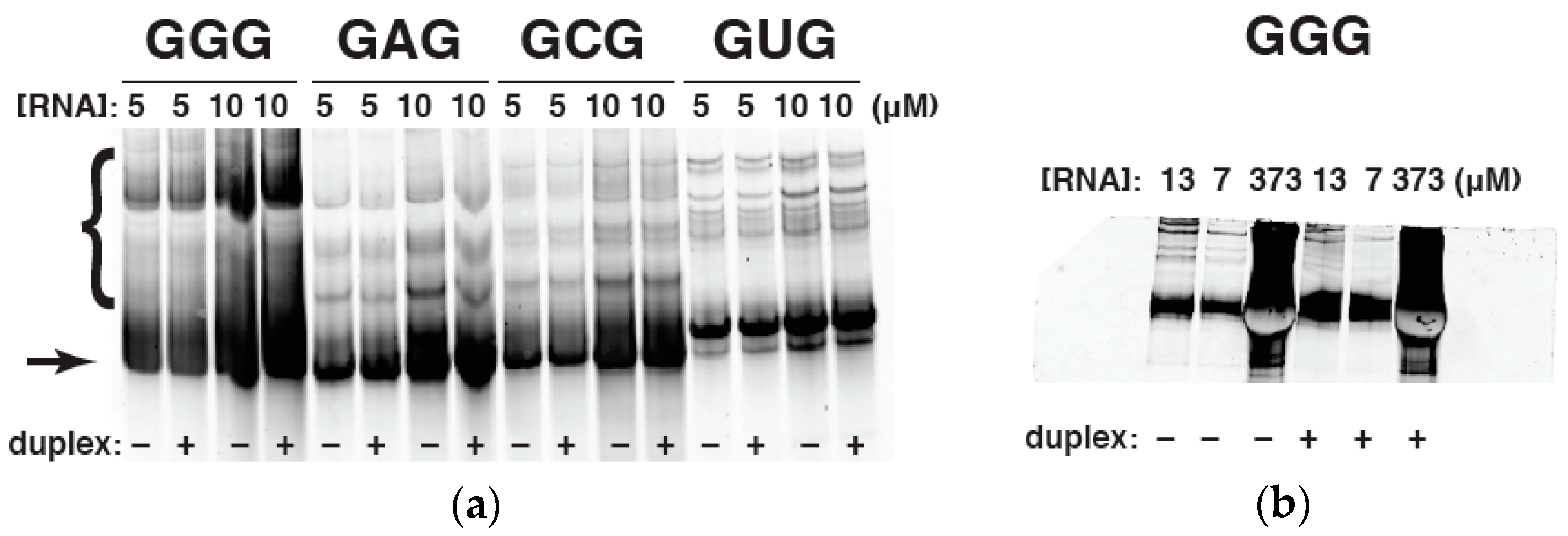
2.4. Native Gel Electrophoresis
3. Discussion
4. Materials and Methods
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A

References
- Joyce, G.F. The antiquity of RNA-based evolution. Nature 2002, 418, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Riley, C.A.; Lehman, N. Generalized RNA-directed recombination of RNA. Chem. Biol. 2003, 10, 1233–1243. [Google Scholar] [CrossRef] [PubMed]
- Hayden, E.J.; Lehman, N. Self-assembly of a group I intron from inactive oligonucleotide fragments. Chem. Biol. 2006, 13, 909–918. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, N.; Manapat, M.L.; Chen, I.A.; Xulvi-Brunet, R.; Hayden, E.J.; Lehman, N. Spontaneous network formation among cooperative RNA replicators. Nature 2012, 491, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Yeates, J.A.; Hilbe, C.; Zwick, M.; Nowak, M.A.; Lehman, N. Dynamics of prebiotic RNA reproduction illuminated by chemical game theory. Proc. Natl. Acad. Sci. USA 2016, 113, 5030–5035. [Google Scholar] [CrossRef] [PubMed]
- Herschlag, D.; Cech, T.R. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 1. Kinetic description of the reaction of an RNA substrate complementary to the active site. Biochemistry 1990, 29, 10159–10171. [Google Scholar] [CrossRef] [PubMed]
- Zaug, A.J.; McEvoy, M.M.; Cech, T.R. Self-splicing of the group I intron from Anabaena pre-tRNA: Requirement for base-pairing of the exons in the anticodon stem. Biochemistry 1993, 32, 7946–7953. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Gooding, A.R.; Cech, T.R. Structure of the Tetrahymena ribozyme: Base triple sandwich and metal ion at the active site. Mol. Cell 2004, 16, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.L.; Stahley, M.R.; Kosek, A.B.; Wang, J.; Strobel, S.A. Crystal structure of a self-splicing group I intron with both exons. Nature 2004, 430, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Karbstein, K.; Lee, J.; Herschlag, D. Probing the role of a secondary structure element at the 5’- and 3′-splice sites in group I intron self-splicing: The Tetrahymena L-16 ScaI ribozyme reveals a new role of the G.U pair in self-splicing. Biochemistry 2007, 46, 4861–4875. [Google Scholar] [CrossRef] [PubMed]
- Kuo, L.Y.; Davidson, L.A.; Pico, S. Characterization of the Azoarcus ribozyme: tight binding to guanosine and substrate by an unusually small group I ribozyme. Biochim. Biophys. Acta 1999, 1489, 281–292. [Google Scholar] [CrossRef]
- Gleitsman, K.R.; Herschlag, D.H. A kinetic and thermodynamic framework for the Azoarcus group I ribozyme reaction. RNA 2014, 20, 1732–1746. [Google Scholar] [CrossRef] [PubMed]
- Dolan, G.F.; Müller, U.F. Trans-splicing with the group I intron ribozyme from Azoarcus. RNA 2014, 20, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Draper, W.E.; Hayden, E.J.; Lehman, N. Mechanisms of covalent self-assembly of the Azoarcus ribozyme from four fragment oligonucleotides. Nucleic Acids Res. 2008, 36, 520–531. [Google Scholar] [CrossRef] [PubMed]
- Denesyuk, N.A.; Thirumalai, D. How do metal ions direct ribozyme folding? Nat. Chem. 2015, 7, 793–801. [Google Scholar] [CrossRef] [PubMed]
- Von Kiedrowski, G. A self-replicating hexadeoxynucleotide. Angew. Chem. Int. Ed. Engl. 1986, 25, 932–935. [Google Scholar] [CrossRef]
- Sievers, D.; von Kiedrowski, G. Self-replication of complementary nucleotide-based oligomers. Nature 1994, 369, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Perrin, F. Polarisation de la lumière de fluorescence: Vie moyenne des molécules dans l’etat excite. J. Phys. Radium 1926, 7, 390–401. [Google Scholar] [CrossRef]
- Narlikar, G.J.; Herschlag, D.H. Isolation of a local tertiary folding transition in the context of a globally folded RNA. Nat. Struct. Mol. Biol. 1996, 3, 701–710. [Google Scholar] [CrossRef]
- Michel, F.; Westhof, E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 1990, 216, 585–610. [Google Scholar] [CrossRef]
- Hayden, E.J.; von Kiedrowski, G.; Lehman, N. Systems chemistry on ribozyme self-construction: Evidence for anabolic autocatalysis in a recombination network. Angew. Chem. Int. Ed. Engl. 2008, 47, 8424–8428. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.H.; Mathews, D.H. NNDB: The nearest neighbor parameter database for predicting stability of nucleic acid secondary structure. Nucleic Acids Res. 2010, 38, D280–D282. [Google Scholar] [CrossRef] [PubMed]
- Leontis, N.B.; Stombaugh, J.; Westhof, E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002, 30, 3497–3531. [Google Scholar] [CrossRef] [PubMed]
- Rangan, P.; Masquida, B.; Westhof, E.; Woodson, S.A. Assembly of core helices and rapid tertiary folding of a small bacterial group I ribozyme. Proc. Natl. Acad. Sci. USA 2003, 100, 1574–1579. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the RNA species in this study are available from the authors.

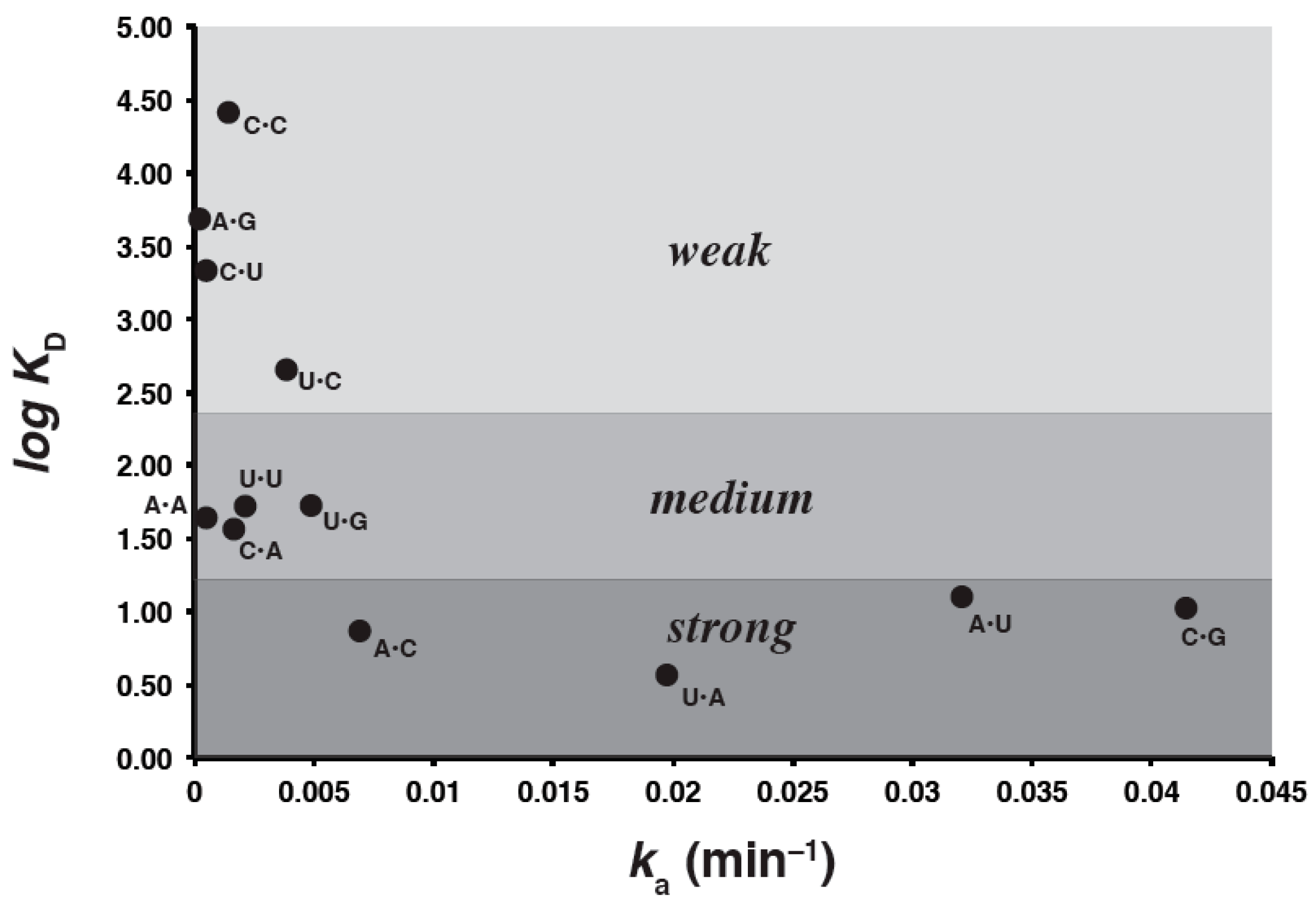
| Genotype a | Self-Assembly Rates (ka) d | ΔG˚48 | KD (μM) from FA Data Herein f |
|---|---|---|---|
| C•G | 0.0415 | –0.89 e | 10.8 |
| A•U | 0.0319 | 0.97 | 12.2 |
| U•A b | 0.0197 | 0.77 | 3.73 |
| G•C | 0.0125 | –1.4 | 1.54 × 103 |
| G•U | 0.0091 | 3.5 | 1.89 × 102 |
| A•C | 0.0069 | 5.5 | 7.62 |
| U•G | 0.0049 | 2.6 | 56.7 |
| U•C | 0.0038 | 5.5 | 4.54 × 102 |
| U•U | 0.0022 | 5.4 | 51.8 |
| C•A | 0.0020 | 4.8 | 39.9 |
| C•C | 0.0016 | 5.5 | 2.62 × 104 |
| G•G | 0.0006 | 2.8 | 1.31 × 104 |
| G•A | 0.0005 | 5.7 | 2.38 × 104 |
| A•A | 0.0004 | 4.9 | 42.4 |
| C•U | 0.0004 | 5.3 | 2.16 × 103 |
| A•G | 0.0001 | 4.0 | 4.79 × 103 |
| G•C c | ND | ND | 63.6 |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Satterwhite, L.E.; Yeates, J.A.M.; Lehman, N. Group I Intron Internal Guide Sequence Binding Strength as a Component of Ribozyme Network Formation. Molecules 2016, 21, 1293. https://doi.org/10.3390/molecules21101293
Satterwhite LE, Yeates JAM, Lehman N. Group I Intron Internal Guide Sequence Binding Strength as a Component of Ribozyme Network Formation. Molecules. 2016; 21(10):1293. https://doi.org/10.3390/molecules21101293
Chicago/Turabian StyleSatterwhite, Laura Elizabeth, Jessica A. M. Yeates, and Niles Lehman. 2016. "Group I Intron Internal Guide Sequence Binding Strength as a Component of Ribozyme Network Formation" Molecules 21, no. 10: 1293. https://doi.org/10.3390/molecules21101293







