Bioactivity of a Novel Glycolipid Produced by a Halophilic Buttiauxella sp. and Improving Submerged Fermentation Using a Response Surface Method
Abstract
:1. Introduction
2. Results
2.1. Bacterial Characterization and Antimicrobial Studies
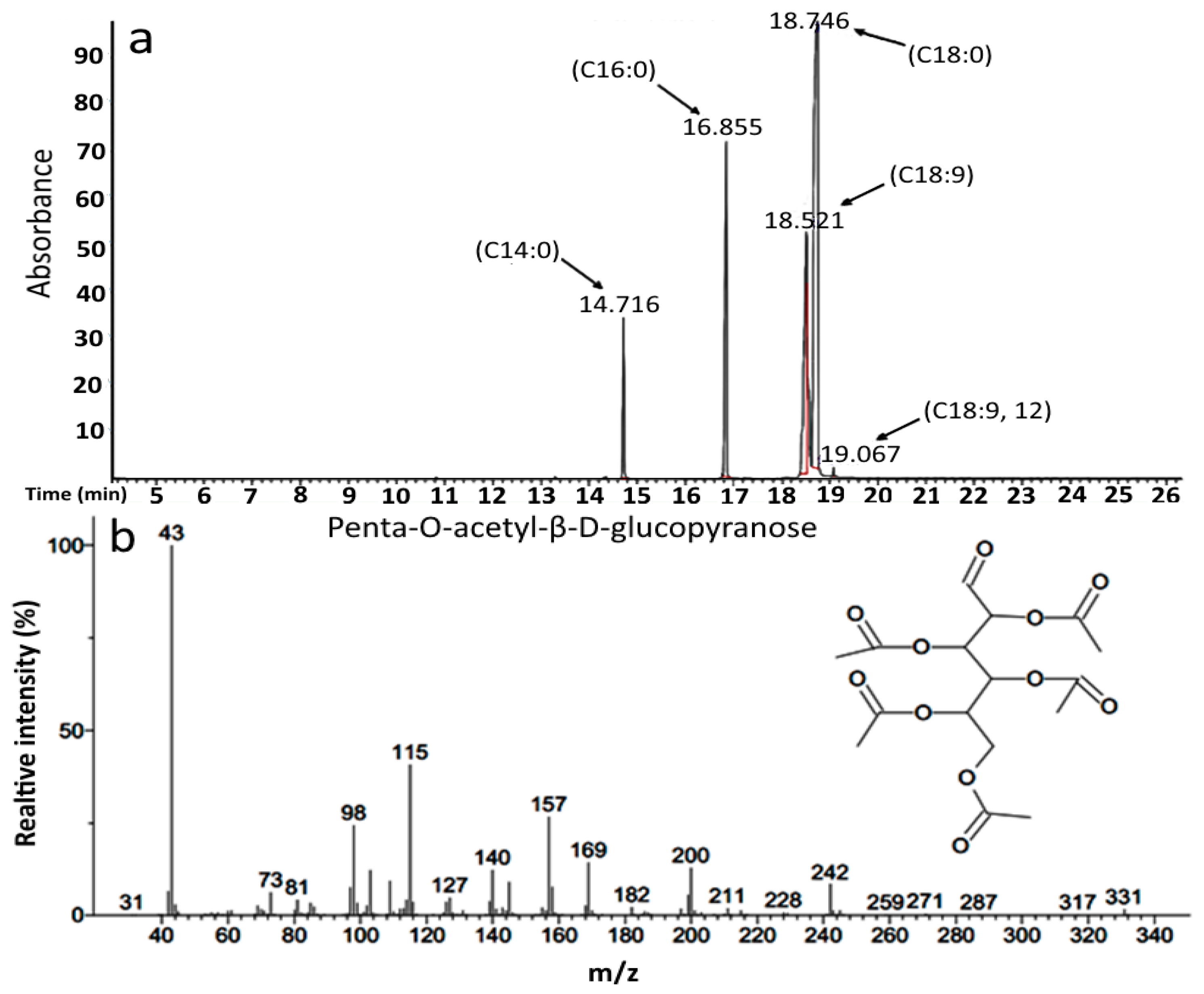
2.2. Compositional Characterization of by GC-MS
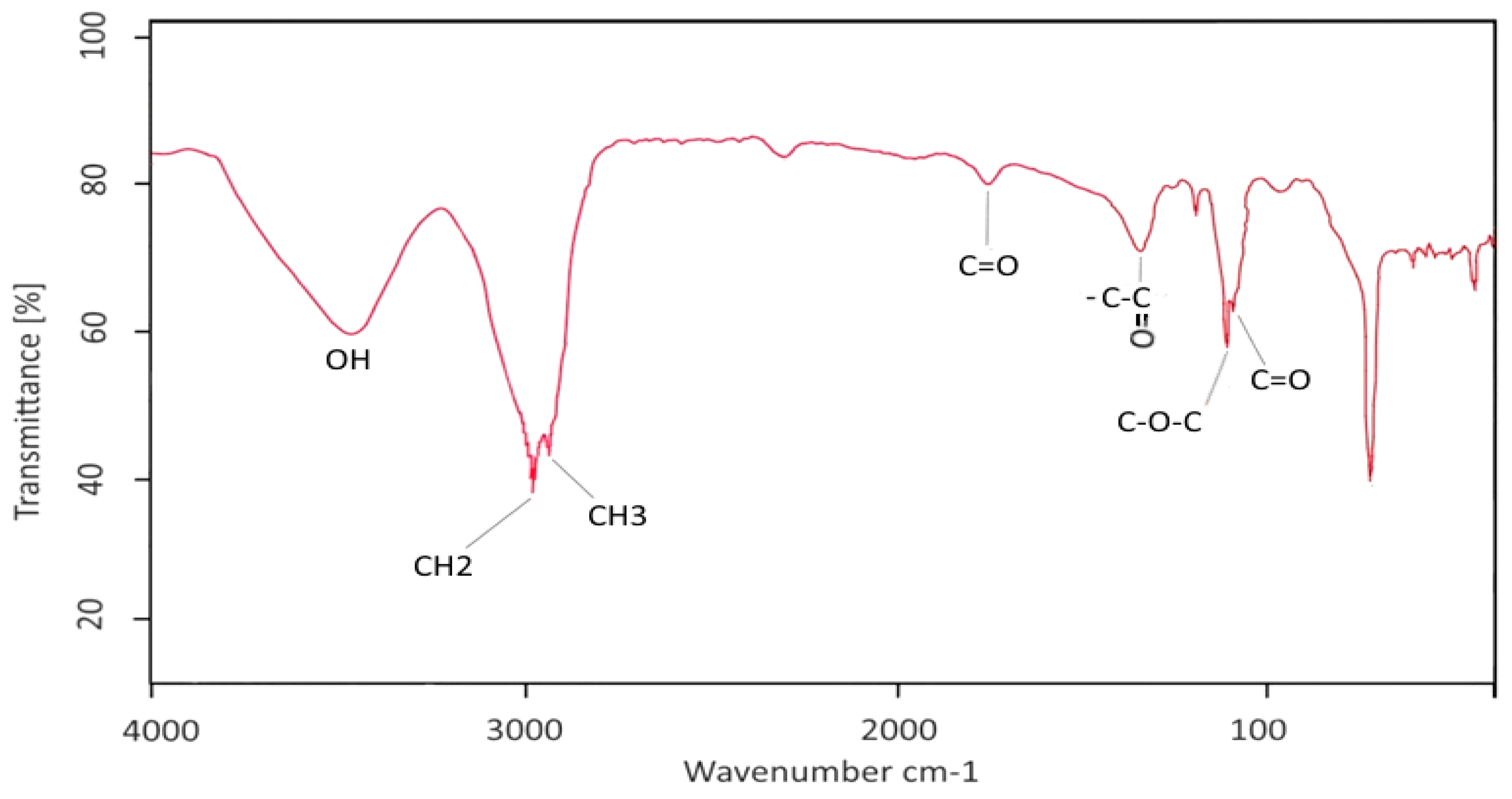
2.3. Characterization of GBS by FT-IR
2.4. Effect of pH, Temperature, and Salinity on GBS Activity
2.5. Optimization of Glycolipid Production by RSM
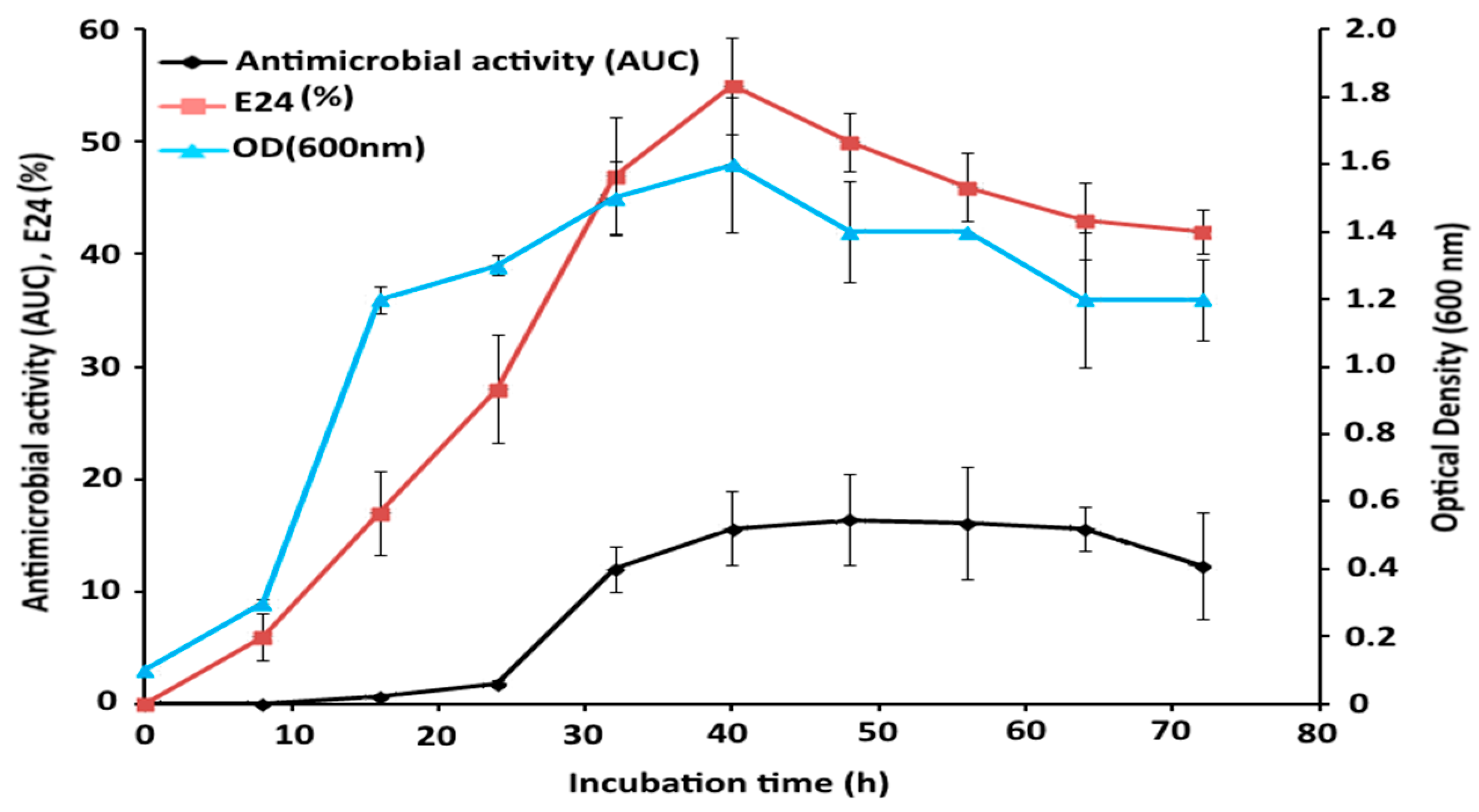
2.6. Time Course Study of Bacterial Growth and GBS Production
3. Discussion
4. Experimental Section
4.1. Bacterial Isolation and Characterization
4.2. Media Preparation and Antimicrobial Activity
4.3. Antimicrobial GBs Purification and Partial Characterization
4.4. Instruments and Analytical Method
4.4.1. Gas Chromatography-Mass Spectrometry (GC-MS)
4.4.2. Fourier Transformed Infrared Spectroscopy (FTIR)
4.4.3. Nuclear Magnetic Resonance Spectroscopy (NMR)
4.5. Physical Stability Studies of the Bioactive Compound
4.6. Optimization of GBS Production by Response Surface Method
4.7. Time-Course Study of GBS Production
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Morya, V.K.; Ahn, C.; Jeon, S.; Kim, E.K. Medicinal and cosmetic potentials of sophorolipids. Mini Rev. Med. Chem. 2013, 13, 1761–1768. [Google Scholar] [CrossRef] [PubMed]
- Barin, R.; Talebi, M.; Biria, D.; Beheshti, M. Fast bioremediation of petroleum-contaminated soils by a consortium of biosurfactant/bioemulsifier producing bacteria. Int. J. Environ. Sci. Technol. 2014, 11, 1701–1710. [Google Scholar] [CrossRef]
- Campos, J.M.; Montenegro Stamford, T.L.; Sarubbo, L.A.; de Luna, J.M.; Rufino, R.D.; Banat, I.M. Microbial biosurfactants as additives for food industries. Biotechnol. Prog. 2013, 29, 1097–1108. [Google Scholar] [CrossRef] [PubMed]
- Gilavand, F.; Marzban, A.; Ebrahimipour, G.; Karkhane, M. Investigation of hydrocarbon bio-removal by the indigenous bacteria isolated from crude oil contaminated soils. J. Microbiol. Biotechnol. Food Sci. 2015, 5, 212–215. [Google Scholar] [CrossRef]
- Deepika, K.V.; Kalam, S.; Sridhar, P.R.; Podile, A.R.; Bramhachari, P.V. Optimization of rhamnolipid biosurfactant production by mangrove sediment bacterium Pseudomonas aeruginosa KVD-HR42 using response surface methodology. Biocatal. Agric. Biotechnol. 2016, 5, 38–47. [Google Scholar] [CrossRef]
- Franzetti, A.; Gandolfi, I.; Bestetti, G.; Smyth, T.J.; Banat, I.M. Production and applications of trehalose lipid biosurfactants. Eur. J. Lipid Sci. Technol. 2010, 112, 617–627. [Google Scholar] [CrossRef]
- Saravanakumari, P.; Mani, K. Structural characterization of a novel xylolipid biosurfactant from Lactococcus lactis and analysis of antibacterial activity against multi-drug resistant pathogens. Bioresour. Technol. 2010, 101, 8851–8854. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Yoo, D.; Kim, Y.; Lee, B.; Shin, D.; Kim, E.K. Characteristics of sophorolipid as an antimicrobial agent. J. Microbiol. Biotechnol. 2002, 12, 235–241. [Google Scholar]
- Ebrahimipour, G.; Gilavand, F.; Karkhane, M.; Kavyanifard, A.; Teymouri, M.; Marzban, A. Bioemulsification activity assessment of an indigenous strain of halotolerant Planococcus and partial characterization of produced biosurfactants. Int. J. Environ. Sci. Technol. 2014, 11, 1379–1386. [Google Scholar] [CrossRef]
- Kaya, K.; Morrison, L.F.; Codd, G.A.; Metcalf, J.S.; Sano, T.; Takagi, H.; Kubo, T. A novel biosurfactant, 2-acyloxyethylphosphonate, isolated from waterblooms of Aphanizomenon flos-aquae. Molecules 2006, 11, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Alit-Susanta, W.G.N.; Takikawa, Y. Phenotypic characterization of Pseudomonas fluorescens PfG32R and its spontaneous gacS mutants and biocontrol activity against bacterial wilt disease of tomato. J. Gen. Plant Pathol. 2006, 72, 168–175. [Google Scholar] [CrossRef]
- Sinanoglou, V.J.; Zoumpoulakis, P.; Heropoulos, G.; Proestos, C.; Ćirić, A.; Petrovic, J.; Glamoclija, J.; Sokovic, M. Lipid and fatty acid profile of the edible fungus Laetiporus sulphurous. Antifungal and antibacterial properties. J. Food Sci. Technol. 2015, 52, 3264–3272. [Google Scholar] [CrossRef] [PubMed]
- Das, P.; Mukherjee, S.; Sen, R. Antimicrobial potential of a lipopeptide biosurfactant derived from a marine Bacillus circulans. J. Appl. Microbiol. 2008, 104, 1675–1684. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, M.; Kagalkar, A.; Jadhav, S.; Govindwar, S. Isolation, characterization, and antifungal application of a biosurfactant produced by Enterobacter sp. MS16. Eur. J. Lipid Sci. Technol. 2011, 113, 1347–1356. [Google Scholar] [CrossRef]
- Fusconi, R.; Maria Nascimento Assunção, R.; de Moura Guimarães, R.; Rodrigues Filho, G.; Eduardo da Hora Machado, A. Exopolysaccharide produced by Gordonia polyisoprenivorans CCT 7137 in GYM commercial medium and sugarcane molasses alternative medium: FT-IR study and emulsifying activity. Carbohydr. Polym. 2010, 79, 403–408. [Google Scholar] [CrossRef]
- Arata, S.; Kurosu, H.; Kuroki, S.; Ando, I. Structural characterization of stearic acid in the crystalline state by the cross polarization in solid state 13C NMR. J. Mol. Struct. 1999, 513, 133–138. [Google Scholar] [CrossRef]
- Gurst, J.E. NMR and the Structure of d-glucose. J. Chem. Educ. 1991, 68, 1003. [Google Scholar] [CrossRef]
- Wei, Y.-H.; Lai, H.-C.; Chen, S.-Y.; Yeh, M.-S.; Chang, J.-S. Biosurfactant production by Serratia marcescens SS-1 and its isogenic strain SMΔR defective in SpnR, a quorum-sensing LuxR family protein. Biotechnol. Lett. 2004, 26, 799–802. [Google Scholar] [CrossRef] [PubMed]
- Dusane, D.H.; Pawar, V.S.; Nancharaiah, Y.V.; Venugopalan, V.P.; Kumar, A.R.; Zinjarde, S.S. Anti-biofilm potential of a glycolipid surfactant produced by a tropical marine strain of Serratia marcescens. Biofouling 2011, 27, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Dalsoo, Y.; Kim, K.; Kim, Y.; Kim, E. Antimicrobial Activity of Biosurfactants. J. Soc. Cosmet. Sci. Korea 2001, 27, 57–58. [Google Scholar]
- Pazzetto, R.; Ferreira, S.B.S.; Santos, E.J.S.; Moriwaki, C.; Guedes, T.A.; Matioli, G. Preservation of Bacillus firmus Strain 37 and Optimization of Cyclodextrin Biosynthesis by Cells Immobilized on Loofa Sponge. Molecules 2012, 17, 9476–9488. [Google Scholar] [CrossRef] [PubMed]
- Zohdi, N.K.; Amid, M. Optimization of Extraction of Novel Pectinase Enzyme Discovered in Red Pitaya (Hylocereus polyrhizus) Peel. Molecules 2013, 18, 14366–14380. [Google Scholar] [CrossRef] [PubMed]
- Shahriari Moghadam, M.; Ebrahimipour, G.; Abtahi, B.; Ghassempour, A.; Hashtroudi, M.S. Biodegradation of polycyclic aromatic hydrocarbons by a bacterial consortium enriched from mangrove sediments. J. Environ. Health Sci. Eng. 2014, 12, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Banat, I.M.; Satpute, S.K.; Cameotra, S.S.; Patil, R.; Nyayanit, N.V. Cost effective technologies and renewable substrates for biosurfactants’ production. Front. Microbiol. 2014, 5, 697. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.-H.; Wang, L.-F.; Changy, J.-S.; Kung, S.-S. Identification of induced acidification in iron-enriched cultures of Bacillus subtilis during biosurfactant fermentation. J. Biosci. Bioeng. 2003, 96, 174–178. [Google Scholar] [CrossRef]
- Joshi, S.; Bharucha, C.; Jha, S.; Yadav, S.; Nerurkar, A.; Desai, A.J. Biosurfactant production using molasses and whey under thermophilic conditions. Bioresour. Technol. 2008, 99, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Ćavar, S.; Maksimović, M.; Vidic, D.; Parić, A. Chemical composition and antioxidant and antimicrobial activity of essential oil of Artemisia annua L. from Bosnia. Ind. Crops Prod. 2012, 37, 479–485. [Google Scholar] [CrossRef]
- Das, P.; Mukherjee, S.; Sen, R. Substrate dependent production of extracellular biosurfactant by a marine bacterium. Bioresour. Technol. 2009, 100, 1015–1019. [Google Scholar] [CrossRef] [PubMed]
- Dubey, K.; Juwarkar, A. Distillery and curd whey wastes as viable alternative sources for biosurfactant production. World J. Microbiol. Biotechnol. 2001, 17, 61–69. [Google Scholar] [CrossRef]
- Gatard, S.; Nasir, M.N.; Deleu, M.; Klai, N.; Legrand, V.; Bouquillon, S. Bolaamphiphiles Derived from Alkenyl l-Rhamnosides and Alkenyl d-Xylosides: Importance of the Hydrophilic Head. Molecules 2013, 18, 6101–6112. [Google Scholar] [CrossRef] [PubMed]
- Dadrasnia, A.; Ismail, S. Biosurfactant production by Bacillus salmalaya for lubricating oil solubilization and biodegradation. Int. J. Environ. Res. Public Health 2015, 12, 9848–9863. [Google Scholar] [CrossRef] [PubMed]
- Anaukwu, C.G.; Ezemba, C.C.; Anakwenze, V.N.; Agu, K.C.; Okeke, B.C.; Awah, N.S.; Ekwealor, I.A. Effect of biosurfactant produced by Citrobacter murliniae AF025369 and a synthetic surfactant on degradation of crude oil. Edorium J. Microbiol. 2016, 2, 1–6. [Google Scholar]
- Velho-Pereira, S.; Kamat, N. Antimicrobial screening of actinobacteria using a modified cross-streak method. Indian J. Pharm. Sci. 2011, 73, 223–228. [Google Scholar] [PubMed]
- Simidu, U.; Hasuo, K. An improved medium for the isolation of bacteria from marine fish. Microbiology 1968, 52, 355–360. [Google Scholar] [CrossRef]
- Ebrahimipour, G.; Moradi, A.; Mehrdad, M.; Marzban, A.; Alaee, H. Evaluation of antimicrobial substance produced by a bacterium isolated from Parmacella iberica. Jundishapur J. Microbiol. 2011, 4, 131–140. [Google Scholar]
- Ichihara, K.I.; Fukubayashi, Y. Preparation of fatty acid methyl esters for gas-liquid chromatography. J. Lipid Res. 2010, 51, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Jerkovic, I.; Mastelic, J. GC-MS Characterization of Acetylatedb-d-glucopyranosides: Transglucosylation of Volatile Alcohols Using Almondb-glucosidase. Croat. Chem. Acta 2004, 77, 529–535. [Google Scholar]
- Metcalfe, L.; Schmitz, A.A.; Pelka, J. Rapid preparation of fatty acid esters from lipids for gas chromatographic analysis. Anal. Chem. 1966, 38, 514–515. [Google Scholar] [CrossRef]
- Sample Availability: Samples of the compounds produced by Buttiexella sp. M44 are available from the authors.

| Test Organism | MIC (µg/mL) | Disc Diffusion (mm) |
|---|---|---|
| E. coli | 200 | 18.5 ± 2.1 |
| S. enterica | 250 | 13.1 ± 3.3 |
| C. albicans | 150 | 23.5 ± 2.3 |
| A. niger | 100 | 26.2 ± 2.7 |
| B. Subtilis | 300 | 14.41 ± 3.2 |
| B. cereus | 250 | 10.60 ± 4.5 |
| S. aureus | 450 | 5.8 ± 1.7 |
| P. aeruginosa | - | - |
| Compound Name | Formula | Molecular Weight |
|---|---|---|
| Tetradecanoic acid methyl ester | C15H30O2 | 242 |
| 9-Hexadecenoic acid methyl ester | C17H32O2 | 268 |
| Hexadecanoic acid methyl ester | C17H34O2 | 270 |
| 9-Octadecenoic acid methyl ester | C19H36O2 | 296 |
| Octadecanoic acid methyl ester | C19H38O2 | 298 |
| 9,12-Octadecadienoic acid methyl ester | C19H34O2 | 294 |
| d-Glucopyranose, 2, 3, 4, 5, 6 pentaacetate | C16H22O11 | 390 |
| Functional Group | d-Glucose | Glycolipid | Octadecanoic Acid | ||||
|---|---|---|---|---|---|---|---|
| 1H-NMR | 13C-NMR | 1H-NMR | 13C-NMR | 1H-NMR | 13C-NMR | ||
| Sugar moiety | C-1, H-1 | 4.92 | 92.6 | 4.55 | 95.1 | - | - |
| C-2, H-2 | 3.53 | 73.5 | 3.13 | 72.8 | - | - | |
| C-3, H-3 | 3.5 | 73.8 | 3.16 | 74.5 | - | - | |
| C-4, H-4 | 3.15 | 70.8 | 3.13 | 69.8 | - | - | |
| C-5, H-5 | 3.53 | 72.6 | 3.58 | 71.5 | - | - | |
| C-6, (6-Hydroxyl) | 1.29 | 61.6 | ** | 65.2 | - | - | |
| Lipid moiety | 1′ (Carboxyl) | - | - | ** | 172.8 * | 11.00 | 180.5 * |
| 2′ (CH2) | - | - | 2.26 | 34.2 | 2.35 | 35.4 | |
| 3′ (CH2) | - | - | 1.56 | 26.3 | 1.64 | 24.7 | |
| -CH2-(C′4-C′16) | - | - | 1.25–1.27 | 29.0–33.1 | 1.26–1.32 | 33.9 | |
| C′17 (CH2) | - | - | 1.23 | 23.7 | 0.93 | 25.1 | |
| C′18 (CH3) | - | - | 0.86 | 14.0 | 0.88 | 15 | |
| Properties | pH | |||
| 4 | 7 | 8 | 12 | |
| ST (mN/m) | 18.4 ± 2.1 | 48.2 ± 3.8 | 50.5 ± 6.4 | 34.7 ± 4.0 |
| OSD (mm) | 10.1 ± 1.7 | 12.4 ± 1.2 | 12.6 ± 1.5 | 11.0 ± 2.2 |
| E24 (%) | 32.1 ± 3.3 | 46.4 ± 4.2 | 48.4 ± 5.4 | 22.7 ± 1.7 |
| Properties | Temperature (°C) | |||
| 20 | 35 | 60 | 100 | |
| ST (mN/m) | 46.6 ± 4.8 | 53.5 ± 6.7 | 50.7 ± 6.5 | 10.0 ± 1.6 |
| OSD (mm) | 10.6 ± 2.5 | 9.7 ± 1.0 | 11.3 ± 2.3 | 12.5 ± 1.2 |
| E24 (%) | 48.3 ± 6.3 | 50.2 ± 7.1 | 54.8 ± 8.5 | 53.9 ± 6.6 |
| Properties | NaCl (%) | |||
| 0 | 3 | 6 | 10 | |
| ST (mN/m) | 51.2 ± 7.6 | 48.4 ± 6.4 | 31.7 ± 5.2 | 28.6 ± 4.3 |
| OSD (mm) | 11.5 ± 1.7 | 12.2 ± 2.7 | 9.5 ± 0.8 | 8.6 ± 1.1 |
| E24 (%) | 44.7 ± 3.8 | 47.2 ± 6.5 | 33.5 ± 1.7 | 24.2 ± 3.0 |
| Run | pH | Temperature (°C) | Peptone (g/L) | NaCl (%) | Molasses (%) | Actual Response | Predicted Response |
|---|---|---|---|---|---|---|---|
| 1 | 7 | 35 | 0.5 | 3 | 1 | 11.34 | 11.08 |
| 2 | 6 | 30 | 0.75 | 4 | 1.5 | 15.06 | 15.75 |
| 3 | 6 | 30 | 1.25 | 2 | 1.5 | 12.67 | 13 |
| 4 | 6 | 20 | 0.75 | 2 | 1.5 | 4.42 | 3.59 |
| 5 | 6 | 30 | 0.75 | 2 | 1.5 | 14.49 | 13.49 |
| 6 | 5 | 25 | 1 | 1 | 1 | 10.4 | 10.78 |
| 7 | 8 | 30 | 0.75 | 2 | 1.5 | 16.39 | 16.45 |
| 8 | 6 | 30 | 0.75 | 2 | 1.5 | 12.17 | 13.49 |
| 9 | 5 | 35 | 1 | 3 | 1 | 8.28 | 7.89 |
| 10 | 6 | 40 | 0.75 | 2 | 1.5 | 0 | 1.02 |
| 11 | 6 | 30 | 0.75 | 0 | 1.5 | 17.07 | 16.57 |
| 12 | 7 | 35 | 1 | 3 | 2 | 10.84 | 10.26 |
| 13 | 5 | 25 | 1 | 3 | 2 | 8.47 | 8.34 |
| 14 | 7 | 35 | 0.5 | 1 | 2 | 16.95 | 16.78 |
| 15 | 6 | 30 | 0.75 | 2 | 1.5 | 14.22 | 13.49 |
| 16 | 6 | 30 | 0.75 | 2 | 0.5 | 15.33 | 15 |
| 17 | 6 | 30 | 0.75 | 2 | 1.5 | 14.05 | 13.49 |
| 18 | 7 | 25 | 0.5 | 3 | 2 | 15.81 | 15.81 |
| 19 | 5 | 25 | 0.5 | 1 | 2 | 8.38 | 8.66 |
| 20 | 6 | 30 | 0.75 | 2 | 1.5 | 13.78 | 13.49 |
| 21 | 7 | 35 | 1 | 1 | 1 | 16.24 | 16.16 |
| 22 | 5 | 35 | 0.5 | 1 | 1 | 8.21 | 8.24 |
| 23 | 5 | 35 | 1 | 1 | 2 | 7.06 | 6.76 |
| 24 | 6 | 30 | 0.25 | 2 | 1.5 | 13.4 | 13.25 |
| 25 | 5 | 35 | 0.5 | 3 | 2 | 6.25 | 5.78 |
| 26 | 4 | 30 | 0.75 | 2 | 1.5 | 6.81 | 6.92 |
| 27 | 7 | 25 | 1 | 1 | 2 | 12.54 | 12.72 |
| 28 | 7 | 25 | 0.5 | 1 | 1 | 9.13 | 9.64 |
| 29 | 6 | 30 | 0.75 | 2 | 1.5 | 12.41 | 13.49 |
| 30 | 6 | 30 | 0.75 | 2 | 2.5 | 13 | 13.51 |
| 31 | 5 | 25 | 0.5 | 3 | 1 | 12.38 | 12.58 |
| 32 | 7 | 25 | 1 | 3 | 1 | 14.6 | 14.69 |
| Source | Terms | p-Value | Source | Terms | p-Value |
| Model | quadratic | <0.0001 * | X2X4 | Interactive | <0.0001 * |
| pH (X1) | Linear | <0.0001 * | X2X5 | Interactive | 0.6633 |
| Temperature (X2) | Linear | 0.0057 * | X3X4 | Interactive | 0.0772 |
| Peptone (X3) | Linear | 0.7495 | X3X5 | Interactive | 0.0008 * |
| NaCl (X4) | Linear | 0.2946 | X4X5 | Interactive | 0.1217 |
| Molasses (X5) | Linear | 0.0730 | X12 | squared | 0.0225 * |
| X1X2 | Interactive | 0.0044 * | X22 | Squared | <0.0001 * |
| X1X3 | Interactive | 0.5965 | X32 | squared | 0.6003 |
| X1X4 | Interactive | 0.3503 | X42 | Squared | 0.0024 * |
| X1X5 | Interactive | 0.0030 * | X52 | squared | 0.2817 |
| X2X3 | Interactive | 0.8642 | Lack of Fit | - | 0.6260 |
| Parameter | Value | Parameter | Value | ||
| Std. Dev. | 0.46 | R-Squared | 0.9814 | ||
| Mean | 5.81 | Adj R-Squared | 0.9475 | ||
| C.V. % | 7.91 | Pred R-Squared | 0.7476 | ||
| PRESS | 31.51 | Adeq Precision | 21.158 | ||
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marzban, A.; Ebrahimipour, G.; Danesh, A. Bioactivity of a Novel Glycolipid Produced by a Halophilic Buttiauxella sp. and Improving Submerged Fermentation Using a Response Surface Method. Molecules 2016, 21, 1256. https://doi.org/10.3390/molecules21101256
Marzban A, Ebrahimipour G, Danesh A. Bioactivity of a Novel Glycolipid Produced by a Halophilic Buttiauxella sp. and Improving Submerged Fermentation Using a Response Surface Method. Molecules. 2016; 21(10):1256. https://doi.org/10.3390/molecules21101256
Chicago/Turabian StyleMarzban, Abdolrazagh, Gholamhossein Ebrahimipour, and Abolghasem Danesh. 2016. "Bioactivity of a Novel Glycolipid Produced by a Halophilic Buttiauxella sp. and Improving Submerged Fermentation Using a Response Surface Method" Molecules 21, no. 10: 1256. https://doi.org/10.3390/molecules21101256