The Biosynthetic Pathways of Tanshinones and Phenolic Acids in Salvia miltiorrhiza
Abstract
:1. Introduction
2. Comparative Transcriptome Provides Informative Data in Biosynthetic Pathway Analysis
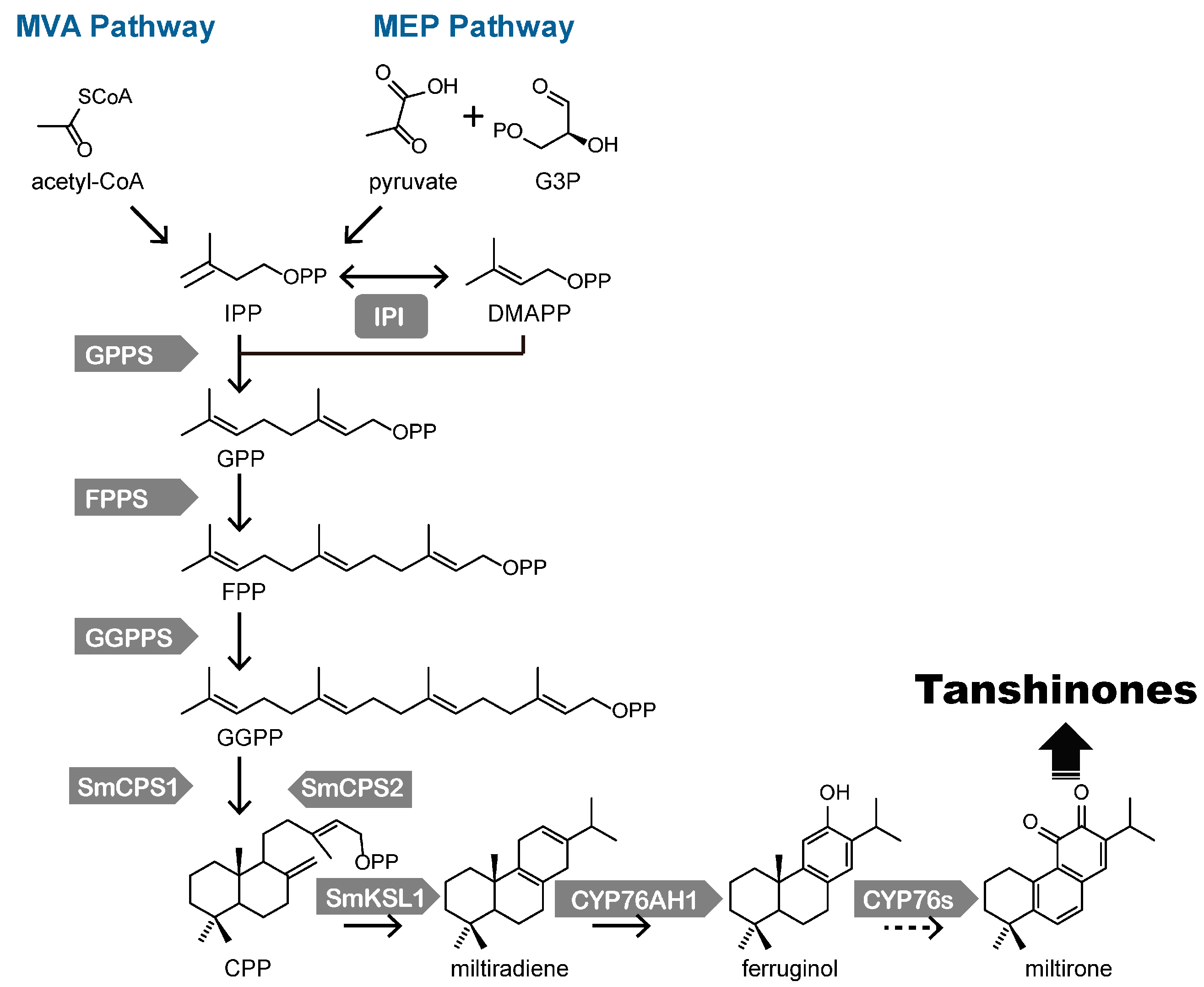
3. Biosynthesis of Tanshinones in S. miltiorrhiza
| Gene (Genbank No.) | Highest Expression Sites | Elicitor | Reference | |
|---|---|---|---|---|
| SmAACT1 (EF635969) | root | YE, Ag+ | [30] | |
| SmAACT2 (AEZ55671) | stem | [31] | ||
| SmHMGS (FJ785326) | leaf | SA, MJ, YE | [32] | |
| SmHMGR1 (EU680958 GU367911) | flower and root | SA, JA, MJ, PEG, ABA | [31,33,34] | |
| SmHMGR2 (FJ747636) | leaf, stem, root | MJ, Ag+ | [22,31,35,36] | |
| SmHMGR3 (JN831102) | leaf | MJ | [31,37] | |
| SmHMGR4 (JN831103) | flower | [31] | ||
| SmMK (JN831104) | stem | MJ | [31] | |
| SmPMK (JN831095) | stem and root | MJ | [31] | |
| SmMDC (JN831105) | stem and root | MJ | [31] | |
| SmDXS1 (EU670744) | leaf | Ag+ | [31,35] | |
| SmDXS2 (FJ643618) | root and leaf | MJ, PEG, ABA, MJ Act12, Ag+ | [17,31,34,35,38] | |
| SmDXS3 (JN831116) | leaf, stem, root cortex, and root stele | MJ | [31] | |
| SmDXS4 (JN831117) | ubiquitous | [31] | ||
| SmDXS5 (JN831118) | leaf and stem | [31] | ||
| SmDXR (FJ768959, FJ476255, DQ991431) | leaf | SA,YE, Hypertonic stress, Act12 PEG, ABA and MJ, Ag+ | [34,35,38,39,40] | |
| SmCMT (JN831096) | leaf | [31] | ||
| SmCMK (EF534309) | leaf | MJ | [31,41] | |
| SmMCS (JX233816) | leaf and stem | Ag+ | [31,42] | |
| SmHDS1 (JN831098) | leaf | [31,43] | ||
| SmHDS2 (KJ746807) | [44] | |||
| SmHDR1 (JX233817 JN831099) | leaf | MJ, Ag+ | [31] | |
| SmHDR2 (JN831100) | leaf | [31] | ||
| SmHDR (JX516088) | leaf | MJ, SA | [45] | |
| SmIPI1 (JN831106) | root | MJ, fungal elicitor | [22,46] | |
| SmIPI2 (JN831106) | stem | MJ | [31] | |
| SmGPPS (JN831107) | [31] | |||
| SmGPPS.LSU (JN831111) | stem and flower | MJ | [31] | |
| SmGPPS.SSUI (JN831108) | leaf | MJ | [31] | |
| SmGPPS.SSUII.1 (JN831109) | leaf and root cortex | [31] | ||
| SmGPPS.SSUII.2 (JN831110) | stem | [31] | ||
| SmFPPS (EF635968 HQ687768) | stem | MJ | [22,31,47] | |
| SmGGPPS1 (FJ643617 FJ178784) | leaf | NaCl, wounding, high temperature, darkness, pathogen, MJ, ABA, SA and GA, Act12, Ag+ | [17,35,38,48,49,50] | |
| SmGGPPS2 (JN831112) | root stele | [31] | ||
| SmGGPPS3 (JN831113) | ubiquitous | [31] | ||
| SmCPS1 (EU003997) | root cortex | Ag+, MJ, YE | [35,51,52] | |
| SmCPS2 (JN831114) | leaf | [31,53] | ||
| SmCPS3 (JN831115) | stem | [31,53] | ||
| SmCPS4 (JN831120) | sepal | [31,53] | ||
| SmCPS5 (JN831121) | stem | MJ | [31,53] | |
| SmKSL1 (EF635966) | root | Ag+, MJ | [35,51,53] | |
| SmKSL2 (JN831119) | root xylem | [31,53] | ||
| CYP76AH1 (JX422213) | root | Ag+ | [27,35] | |
| CYP76s (KR140168) (KR140169) | root | Ag+ | [54] | |
| SmCPR1(FR693803) | [27,55] | |||
| SmCPR2(JX848592) | [27] |
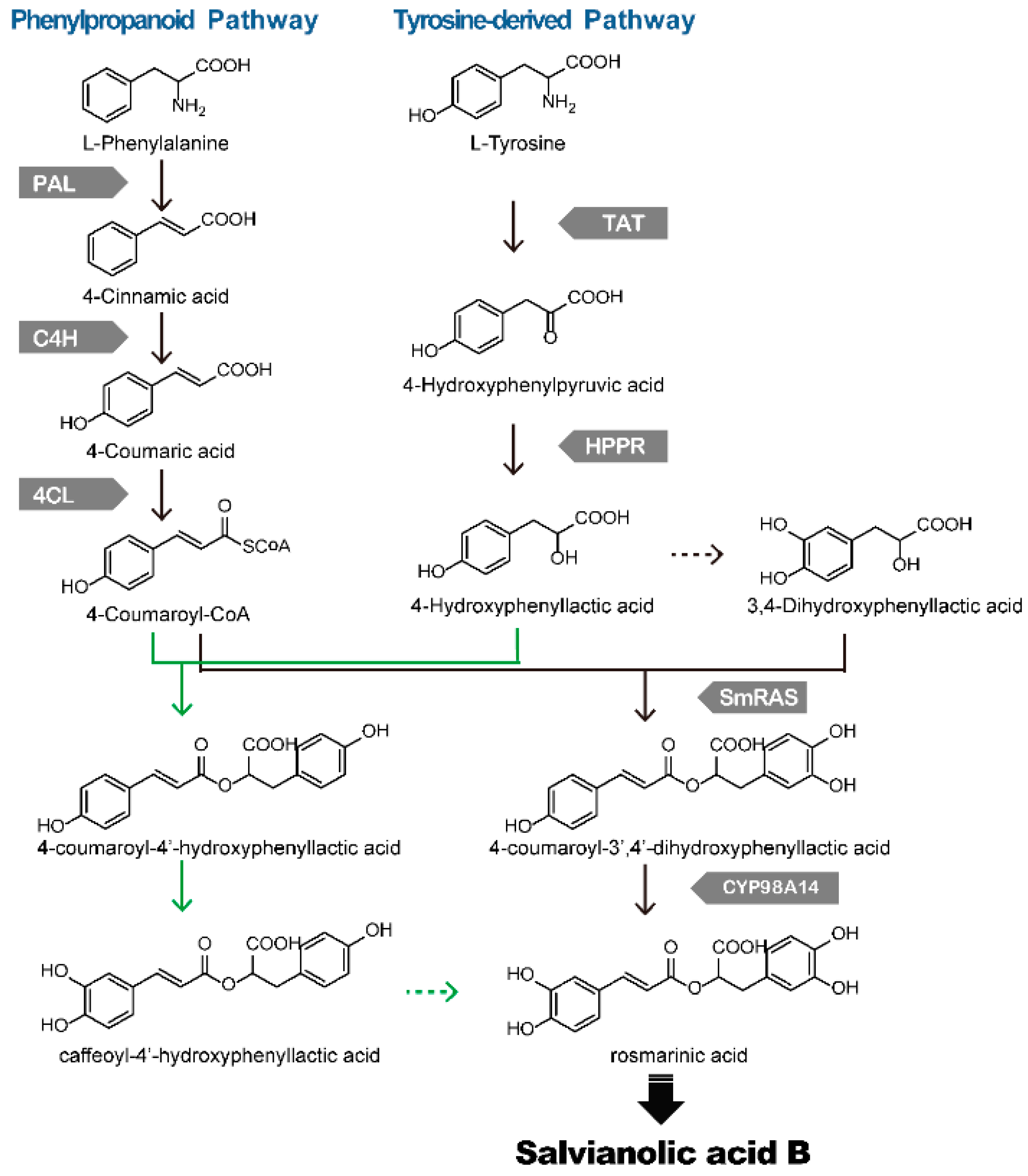
4. Biosynthesis of Phenolic Acids in S. miltiorrhiza
| Gene (Genbank No.) | Highest Expression Sites | Elicitor | Reference |
|---|---|---|---|
| SmPAL1 (EF462460) | root, leaf | ABA, wounding and dehydration, PEG, MJ, SA, Ca2+, GA, ethylene | [66,67,68,71,72,73,74,75] |
| SmPAL2 (GQ249111) | stem, flower | PEG and MJ | [66,68,71] |
| SmPAL3 (KF220569) | rootleaf | PEG and MJ | [66,71] |
| SmC4H1 (EF377337 DQ355979) | root, stem | MJ, ABA, UV-B, Ag+ | [35,71,76,77] |
| SmC4H2 (KF220564) | stem, root | [71] | |
| Sm4CL1 (AY237163) | leaf | MJ, YE | [69,70] |
| Sm4CL2 (AY237164) | root | MJ, YE, Ag+ | [35,69,70,71] |
| Sm4CL3 (KF220556) | root | MJ | [71] |
| Sm4CL-like1 (KF220557) | root | MJ | [71] |
| Sm4CL-like2 (KF220558) | MJ | [71] | |
| Sm4CL-like3 (KF220559) | [71] | ||
| Sm4CL-like4 (KF220560) | root | [71] | |
| Sm4CL-like5 (KF220561) | [71] | ||
| Sm4CL-like6 (KF220562) | MJ | [71] | |
| Sm4CL-like7 (KF220563) | [71] | ||
| SmTAT1 (DQ334606 EF192320) | stem | MJ, ABA, SA, UV-B, GA, ethylene, Ag+, YE | [35,65,72,74,78] |
| SmTAT2 (KF220575) | flower | MJ | [71] |
| SmTAT3 (KF220555) | Stem, root | [71] | |
| SmHPPR1 (DQ099741 DQ266514 EF458148) | stemflower | MJ, SA, GA3, ABA, UV-B, Ag+ | [35,71,79] |
| SmHPPR2 (KF220565) | Stem,leaf | [71] | |
| SmHPPR3 (KF220566) | stem | [71] | |
| SmHPPR4 (KF220567) | [80] | ||
| SmRAS-Like (GU647199) | stem | Pseudomonas lachrymans, MJ, light, and SA | [81] |
| SmRAS1 (FJ906696) | root | MJ, Ag+ | [15,35] |
| SmHCT1 (KF220570) | root | [71] | |
| SmHCT2 (KF220571) | stem | [71] | |
| SmHCT3 (KF220572) | stem | MJ | [71] |
| SmHCT4 (KF220573) | stem | MJ | [71] |
| SmHCT5 (KF220574) | stem | MJ | [71] |
| CYP98A14 (HQ316179) | root | MJ, Ag+ | [15,35] |
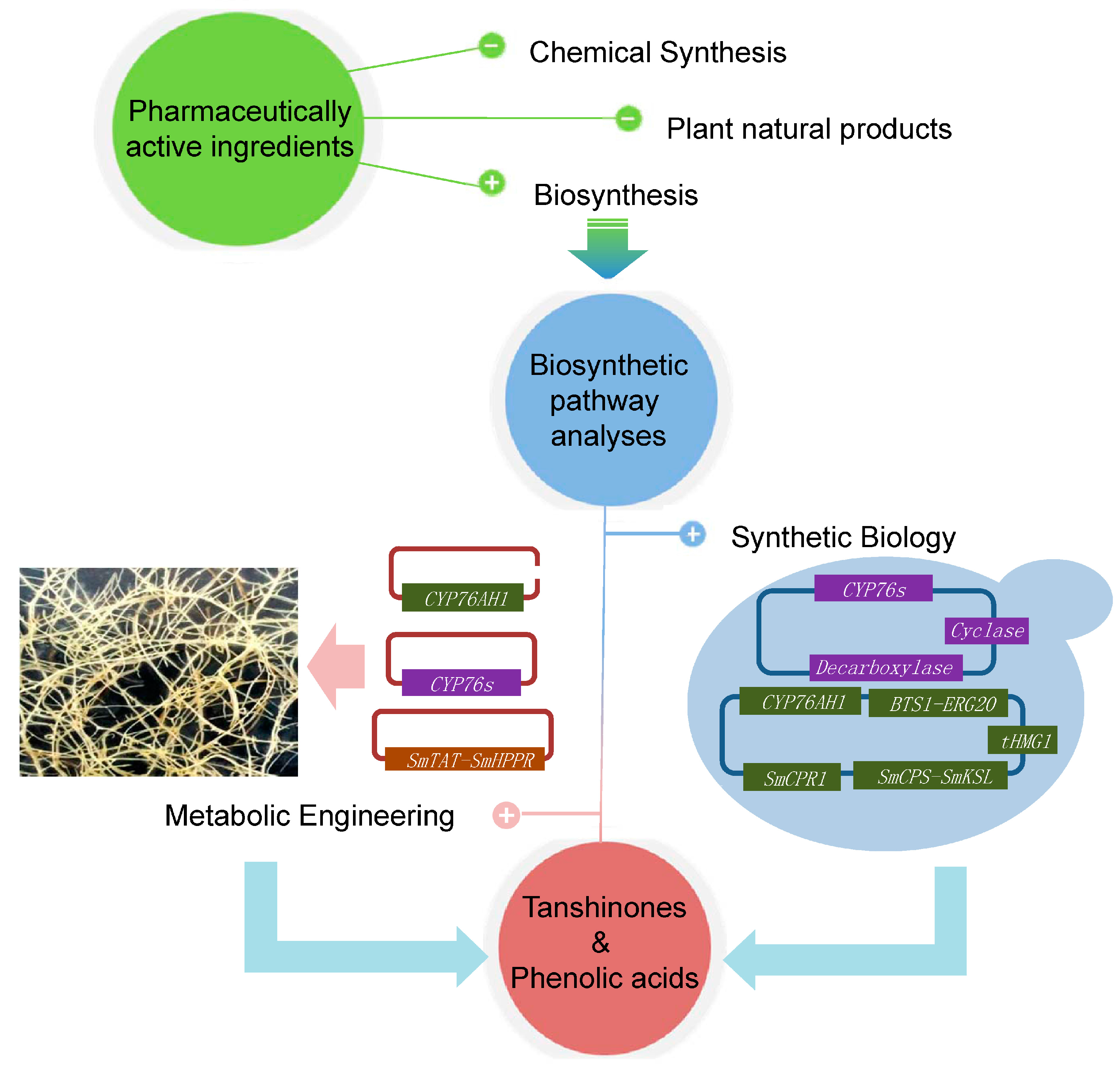
5. Biotechnological Applications
6. Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Croteau, R.; Ketchum, R.B.; Long, R.; Kaspera, R.; Wildung, M. Taxol biosynthesis and molecular genetics. Phytochem. Rev. 2006, 5, 75–97. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Gao, C.; Cui, Z. Ginkgolide a reduces inflammatory response in high-glucose-stimulated human umbilical vein endothelial cells through STAT3-mediated pathway. Int. Immunopharmacol. 2015, 25, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ding, R.; Chai, Y.; Bonfill, M.; Moyano, E.; Oksman-Caldentey, K.M.; Xu, T.; Pi, Y.; Wang, Z.; Zhang, H.; et al. Engineering tropane biosynthetic pathway in Hyoscyamus niger hairy root cultures. Proc. Natl. Acad. Sci. USA 2004, 101, 6786–6791. [Google Scholar] [CrossRef] [PubMed]
- Han, J.Y.; Kim, M.J.; Ban, Y.W.; Hwang, H.S.; Choi, Y.E. The involvement of β-Amyrin 28-oxidase (CYP716A52v2) in oleanane-type ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 2013, 54, 2034–2046. [Google Scholar] [CrossRef] [PubMed]
- Paddon, C.J.; Westfall, P.J.; Pitera, D.J.; Benjamin, K.; Fisher, K.; McPhee, D.; Leavell, M.D.; Tai, A.; Main, A.; Eng, D.; et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 2013, 496, 528–532. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Morris-Natschke, S.L.; Lee, K.-H. Biosynthesis, total syntheses, and antitumor activity of tanshinones and their analogs as potential therapeutic agents. Nat. Prod. Rep. 2011, 28, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Li, M.H.; Peng, Y.; Xiao, P.G. Distribution of tanshinones in the genus Salvia (family Lamiaceae) from China and its systematic significance. J. Syst. Evol. 2010, 48, 118–122. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiang, P.; Ye, M.; Kim, S.H.; Jiang, C.; Lu, J. Tanshinones: Sources, pharmacokinetics and anti-cancer activities. Int. J. Mol. Sci. 2012, 13, 13621–13666. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Guo, J.; Bao, J.; Lu, J.; Wang, Y. The anticancer properties of Salvia miltiorrhiza Bunge (Danshen): A systematic review. Med. Res. Rev. 2014, 34, 768–794. [Google Scholar] [CrossRef] [PubMed]
- Robertson, A.L.; Holmes, G.R.; Bojarczuk, A.N.; Burgon, J.; Loynes, C.A.; Chimen, M.; Sawtell, A.K.; Hamza, B.; Willson, J.; Walmsley, S.R.; et al. A zebrafish compound screen reveals modulation of neutrophil reverse migration as an anti-inflammatory mechanism. Sci. Transl. Med. 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.W.; Zeng, L.H.; Fung, K.P.; Wu, J.; Pang, H.; Grey, A.A.; Weisel, R.D.; Wang, J.Y. Effect of sodium tanshinone IIA sulfonate in the rabbit myocardium and on human cardiomyocytes and vascular endothelial cells. Biochem. Pharmacol. 1993, 46, 2327–2332. [Google Scholar] [CrossRef]
- Wu, Z.; Song, L.; Liu, S.; Huang, D. Tanshinones extend chronological lifespan in budding yeast Sacchraromyces cerevisiae. Appl. Microbiol. Biotechnol. 2014, 98, 8617–8628. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M. Rosmarinic acid: New aspects. Phytochem. Rev. 2013, 12, 207–227. [Google Scholar] [CrossRef]
- Wang, X.; Morris-Natschke, S.L.; Lee, K.-H. New developments in the chemistry and biology of the bioactive constituents of tanshen. Med. Res. Rev. 2007, 27, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Di, P.; Zhang, L.; Chen, J.; Tan, H.; Xiao, Y.; Dong, X.; Zhou, X.; Chen, W. 13C Tracer reveals phenolic acids biosynthesis in hairy root cultures of Salvia miltiorrhiza. ACS Chem. Biol. 2013, 8, 1537–1548. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Su, P.; Hu, Y.; He, Y.; Gao, W.; Huang, L. RNA interference-mediated repression of SmCPS (copalyldiphosphate synthase) expression in hairy roots of Salvia miltiorrhiza causes a decrease of tanshinones and sheds light on the functional role of SmCPS. Biotechnol. Lett. 2014, 36, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Kai, G.; Xu, H.; Zhou, C.; Liao, P.; Xiao, J.; Luo, X.; You, L.; Zhang, L. Metabolic engineering tanshinone biosynthetic pathway in Salvia miltiorrhiza hairy root cultures. Metab. Eng. 2011, 13, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Zhang, L.; Gao, S.; Saechao, S.; Di, P.; Chen, J.; Chen, W. The c4h, tat, hppr and hppd genes prompted engineering of rosmarinic acid biosynthetic pathway in salvia miltiorrhiza hairy root cultures. PLoS ONE 2011, 6, e29713. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Chen, F.; Zhang, Y.L.; Song, J.Y. Production of lithospermic acid B and rosmarinic acid in hairy root cultures of Salvia miltiorrhiza. J. Ind. Microbiol. Biotechnol. 1999, 22, 133–138. [Google Scholar] [CrossRef]
- Hu, Z.; Alfermann, A.W. Diterpenoid production in hairy root cultures of Salvia miltiorrhiza. Phytochemistry 1993, 32, 699–703. [Google Scholar]
- Yuan, Y.; liu, Y.; Lu, D.; Huang, L.; Liang, R.; Yang, Z.; Chen, S. Genetic stability, active constituent, and pharmacoactivity of Salvia miltiorrhiza hairy roots and wild plant. Z. Naturforsch. C. 2009, 64, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Cui, G.; Huang, L.; Tang, X.; Zhao, J. Candidate genes involved in tanshinone biosynthesis in hairy roots of Salvia miltiorrhiza revealed by cDNA microarray. Mol. Biol. Rep. 2011, 38, 2471–2478. [Google Scholar] [CrossRef] [PubMed]
- Hua, W.; Zhang, Y.; Song, J.; Zhao, L.; Wang, Z. De novo transcriptome sequencing in Salvia miltiorrhiza to identify genes involved in the biosynthesis of active ingredients. Genomics 2011, 98, 272–279. [Google Scholar]
- Yang, L.; Ding, G.; Lin, H.; Cheng, H.; Kong, Y.; Wei, Y.; Fang, X.; Liu, R.; Wang, L.; Chen, X.; et al. Transcriptome analysis of medicinal plant Salvia miltiorrhiza and identification of genes related to tanshinone biosynthesis. PLoS ONE 2013, 8, e80464. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Zhu, Y.; Song, J.; Xu, L.; Sun, C.; Zhang, X.; Xu, Y.; He, L.; Sun, W.; Xu, H.; et al. Transcriptional data mining of Salvia miltiorrhiza in response to methyl jasmonate to examine the mechanism of bioactive compound biosynthesis and regulation. Physiol. Plant. 2014, 152, 241–255. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Sun, H.-X.; Xiao, H.; Cui, G.; Hillwig, M.; Jackson, A.; Wang, X.; Shen, Y.; Zhao, N.; Zhang, L.; et al. Combining metabolomics and transcriptomics to characterize tanshinone biosynthesis in Salvia miltiorrhiza. BMC Genomics 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zhou, Y.J.; Hillwig, M.L.; Shen, Y.; Yang, L.; Wang, Y.; Zhang, X.; Liu, W.; Peters, R.J.; Chen, X.; et al. CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proc. Natl. Acad. Sci. USA 2013, 110, 12108–12113. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wu, B.; Nelson, D.R.; Wu, K.; Liu, C. Computational identification and systematic classification of novel cytochrome P450 genes in Salvia miltiorrhiza. PLoS ONE 2015, 10, e0120063. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Peters, R.J.; Weirather, J.; Luo, H.; Liao, B.; Zhang, X.; Zhu, Y.; Ji, A.; Zhang, B.; Hu, S.; et al. Full-length transcriptome sequences and splice variants obtained by a combination of sequencing platforms applied to different root tissues of Salvia miltiorrhiza and tanshinone biosynthesis. Plant. J. 2015, 82, 951–961. [Google Scholar] [CrossRef] [PubMed]
- Cui, G.H.; Wang, X.Y.; Feng, H.; Zhao, J.X.; Huang, L.Q. Molecular cloning and SNP analysis of a acetyl-CoA C-acetyltransferase gene (SmAACT) from Salvia miltiorrhiza. Acta Pharm. Sin. 2010, 45, 785–790. [Google Scholar]
- Ma, Y.; Yuan, L.; Wu, B.; Li, X.; Chen, S.; Lu, S. Genome-wide identification and characterization of novel genes involved in terpenoid biosynthesis in Salvia miltiorrhiza. J. Exp. Bot. 2012, 63, 2809–2823. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yan, X.; Wang, J.; Li, S.; Liao, P.; Kai, G. Molecular cloning and expression analysis of a new putative gene encoding 3-hydroxy-3-methylglutaryl-CoA synthase from Salvia miltiorrhiza. Acta Physiol. Plant. 2011, 33, 953–961. [Google Scholar] [CrossRef]
- Liao, P.; Zhou, W.; Zhang, L.; Wang, J.; Yan, X.; Zhang, Y.; Zhang, R.; Li, L.; Zhou, G.; Kai, G. Molecular cloning, characterization and expression analysis of a new gene encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase from Salvia miltiorrhiza. Acta Physiol. Plant. 2009, 31, 565–572. [Google Scholar] [CrossRef]
- Yang, D.; Ma, P.; Liang, X.; Wei, Z.; Liang, Z.; Liu, Y.; Liu, F. PEG and ABA trigger methyl jasmonate accumulation to induce the MEP pathway and increase tanshinone production in Salvia miltiorrhiza hairy roots. Physiol. Plant. 2012, 146, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Xing, B.; Yang, D.; Guo, W.; Liang, Z.; Yan, X.; Zhu, Y.; Liu, Y. Ag+ as a more effective elicitor for production of tanshinones than phenolic acids in Salvia miltiorrhiza hairy roots. Molecules 2014, 20, 309–324. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Cui, G.; Zhou, S.-F.; Zhang, X.; Huang, L. Cloning and characterization of a novel 3-hydroxy-3-methylglutaryl coenzyme A reductase gene from Salvia miltiorrhiza involved in diterpenoid tanshinone accumulation. J. Plant Physiol. 2011, 168, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Guo, J.; Shen, Y.; Huang, L. Cloning and expression analysis of a new 3-hydroxy-3-methylglutaryl coenzyme A reductase gene from Salvia miltiorrhiza (SmHMGR3). China J. Chin. Mater. Med. 2012, 37, 2378–2382. [Google Scholar]
- Yan, Y.; Zhang, S.; Yang, D.; Zhang, J.; Liang, Z. Effects of Streptomyces pactum Act12 on Salvia miltiorrhiza hairy root growth and tanshinone synthesis and its mechanisms. Appl. Biochem. Biotechnol. 2014, 173, 883–893. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.J.; Shi, M.; Wu, J.Y. Cloning and characterization of the 1-deoxy-d-xylulose 5-phosphate reductoisomerase gene for diterpenoid tanshinone biosynthesis in Salvia miltiorrhiza (Chinese sage) hairy roots. Biotechnol. Appl. Biochem. 2009, 52, 89. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Zhang, L.; Wang, J.; Liao, P.; Zhang, Y.; Zhang, R.; Kai, G. Molecular characterization and expression of 1-deoxy-d-xylulose 5-phosphate reductoisomerase (DXR) gene from Salvia miltiorrhiza. Acta Physiol. Plant. 2009, 31, 1015–1022. [Google Scholar] [CrossRef]
- Wang, X.Y.; Cui, G.H.; Huang, L.Q.; Gao, W.; Yuan, Y. A full length cDNA of 4-(cytidine 5′-diphospho)-2-C-methyl-d-erythritol kinase cloning and analysis of introduced gene expression in Salvia miltiorrhiza. Acta Pharm. Sin. 2008, 43, 1251–1257. [Google Scholar]
- Gao, W.; Cheng, Q.; Ma, X.; He, Y.; Jiang, C.; Yuan, Y.; Huang, L. Cloning and bioinformatics analysis of 2-C-methyl-d-erythritol 2,4-cyclodiphosphate synthase gene in Salvia miltiorrhiza. China J. Chin. Mater. Med. 2012, 37, 3365–3370. [Google Scholar]
- Cheng, Q.Q.; He, Y.F.; Li, G.; Jiang, C.; Yuan, Y.; Gao, W.; Huang, L.Q. Cloning and induced expression analysis of 4-hydroxy-3-methyl-but-2-enyl diphosphate reductase gene (SmHDR) of Salvia miltiorrhiza. Acta Pharm. Sin. 2013, 48, 236–242. [Google Scholar]
- Jiang, D.; Rong, Q.; Yuan, Q.; Zhang, W.; Zhang, Y.; Huang, L. Cloning and prokaryotic expression analysis of HDS from Salvia miltiorrhiza bge.f.alba. Acta Pharm. Sin. 2014, 49, 1614–1620. [Google Scholar]
- Hao, G.; Shi, R.; Tao, R.; Fang, Q.; Jiang, X.; Ji, H.; Feng, L.; Huang, L. Cloning, molecular characterization and functional analysis of 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate reductase (HDR) gene for diterpenoid tanshinone biosynthesis in Salvia miltiorrhiza Bge. f. alba. Plant Physiol. Biochem. 2013, 70, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhou, L.; Hua, W.; Wang, Z.; Li, C. Bioinformatics and expression analysis of isopentenyl diphosphate isomerase (SmIPI) from Salvia militiorrhiza Bunge. Plant Phisiol. J. 2011, 47, 1086–1090. [Google Scholar]
- Zhou, L.; Hua, W.; Yang, Y.; Wang, Z.; Li, C. Expression pattern of farnesyl diphosphate synthese (SmFPPS1) from Salvia Miltiorrhiza Bunge. J. Shaanxi Norm. Univ. (Nat. Sci. Ed.) 2013, 41, 70–75. [Google Scholar]
- Hua, W.; Song, J.; Li, C.; Wang, Z. Molecular cloning and characterization of the promoter of SmGGPPs and its expression pattern in Salvia miltiorrhiza. Mol. Biol. Rep. 2012, 39, 5775–5783. [Google Scholar] [CrossRef] [PubMed]
- Kai, G.; Liao, P.; Zhang, T.; Zhou, W.; Wang, J.; Xu, H.; Liu, Y.; Zhang, L. Characterization, expression profiling, and functional identification of a gene encoding geranylgeranyl diphosphate synthase from Salvia miltiorrhiza. Biotechnol. Bioprocess Eng. 2010, 15, 236–245. [Google Scholar] [CrossRef]
- Zhang, L.; Dai, Z.; Cui, G.; Cheng, Y.; Qi, X.; Gao, Z. Cloning and characterization of geranylgeranyl diphosphate synthase gene of Salvia miltiorrhia. China J. Chin. Mater. Med. 2009, 34, 2704–2708. [Google Scholar]
- Gao, W.; Hillwig, M.L.; Huang, L.; Cui, G.; Wang, X.; Kong, J.; Yang, B.; Peters, R.J. A functional genomics approach to tanshinone biosynthesis provides stereochemical insights. Org. Lett. 2009, 11, 5170–5173. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; He, Y.; Li, G.; Liu, Y.; Gao, W.; Huang, L. Effects of combined elicitors on tanshinone metabolic profiling and SmCPS expression in Salvia miltiorrhiza hairy root cultures. Molecules 2013, 18, 7473–7485. [Google Scholar] [CrossRef] [PubMed]
- Cui, G.; Duan, L.; Jin, B.; Qian, J.; Xue, Z.; Shen, G.; Snyder, J.H.; Song, J.; Chen, S.; Huang, L.; et al. Functional divergence of diterpene syntheses in the medicinal plant Salvia miltiorrhiza Bunge. Plant. Physiol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Ma, X.H.; Cai, Y.; Ma, Y.; Zhan, Z.L.; Zhou, Y.J.; Liu, W.J.; Guan, M.X.; Cui, G.H.; Kang, L.P.; et al. Cytochrome P450 Promiscuity Leads to a Bifurcating Pathway in Tanshinone Biosynthesis; National Resource Center for Chinese Materia Medica, China Academy of Chinese Medical Sciences: Beijing, China, 2015; Unpublished work. [Google Scholar]
- Cai, Y.; Guo, J.; Zhou, Y.; Zhu, Z.; Wu, W.; Huang, L.; Chen, M.; Zhao, Z. Construction of Saccharomyces cerevisiae whole-cell biocatalyst system for conversion miltiradiene. Acta Pharm. Sin. 2013, 48, 1618–1623. [Google Scholar]
- Rohmer, M.; Knani, M.; Simonin, P.; Sutter, B.; Sahm, H. Isoprenoid biosynthesis in bacteria: A novel pathway for the early steps leading to isopentenyl diphosphate. Biochem. J. 1993, 295, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, M.K. Terpen-Biosynthese in Ginkgo biloba: Eine Überraschende Geschichte. Ph.D. Thesis, ETH Zurich, Zurich, Switzerland, 1994. [Google Scholar]
- Rodrı́guez-Concepción, M.; Boronat, A. Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol. 2002, 130, 1079–1089. [Google Scholar] [CrossRef] [PubMed]
- Miziorko, H.M. Enzymes of the mevalonate pathway of isoprenoid biosynthesis. Arch. Biochem. Biophys. 2011, 505, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Wu, J. Tanshinone production and isoprenoid pathways in Salvia miltiorrhiza hairy roots induced by Ag+ and yeast elicitor. Plant Sci. 2005, 168, 487–491. [Google Scholar] [CrossRef]
- Yang, D.; Du, X.; Liang, X.; Han, R.; Liang, Z.; Liu, Y.; Liu, F.; Zhao, J. Different roles of the mevalonate and methylerythritol phosphate pathways in cell growth and tanshinone production of Salvia miltiorrhiza hairy roots. PLoS ONE 2012, 7, e46797. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Luo, X.; Ju, G.; Yu, X.; Hao, X.; Huang, Q.; Xiao, J.; Cui, L.; Kai, G. Increased accumulation of the cardio-cerebrovascular disease treatment drug tanshinone in Salvia miltiorrhiza hairy roots by the enzymes 3-hydroxy-3-methylglutaryl CoA reductase and 1-deoxy-d-xylulose 5-phosphate reductoisomerase. Funct. Integr. Genomics 2014, 14, 603–615. [Google Scholar] [CrossRef] [PubMed]
- Kirby, J.; Keasling, J.D. Biosynthesis of plant isoprenoids: Perspectives for microbial engineering. Annu. Rev. Plant Biol. 2009, 60, 335–355. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yan, Y.; Wang, B.; Liang, Z.; Liu, Y.; Liu, F.; Qi, Z. Selective responses of enzymes in the two parallel pathways of rosmarinic acid biosynthetic pathway to elicitors in Salvia miltiorrhiza hairy root cultures. J. Biosci. Bioeng. 2014, 117, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Shi, M.; Ng, J.; Wu, J.Y. Elicitor-induced rosmarinic acid accumulation and secondary metabolism enzyme activities in Salvia miltiorrhiza hairy roots. Plant Sci. 2006, 170, 853–858. [Google Scholar] [CrossRef]
- Hou, X.; Shao, F.; Ma, Y.; Lu, S. The phenylalanine ammonia-lyase gene family in Salvia miltiorrhiza: Genome-wide characterization, molecular cloning and expression analysis. Mol. Biol. Rep. 2013, 40, 4301–4310. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Wang, Z. Molecular cloning, expression and characterization of a phenylalanine ammonia-lyase gene (SmPAL1) from Salvia miltiorrhiza. Mol. Biol. Rep. 2009, 36, 939–952. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Wang, Z. RNAi-mediated suppression of the phenylalanine ammonia-lyase gene in Salvia miltiorrhiza causes abnormal phenotypes and a reduction in rosmarinic acid biosynthesis. J. Plant Res. 2011, 124, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.Q.; Chen, Z.W.; Tan, R.H.; Zhao, S.J.; Hu, Z.B. Isolation and functional analysis of 4-coumarate: Coenzyme A ligase gene promoters from Salvia miltiorrhiza. Biol. Plant 2012, 56, 261–268. [Google Scholar] [CrossRef]
- Zhao, S.-J.; Hu, Z.-B.; Liu, D.; Leung, F.C.C. Two divergent members of 4-coumarate: Coenzyme A ligase from Salvia miltiorrhiza Bunge: cDNA cloning and functional study. J. Integr. Plant Biol. 2006, 48, 1355–1364. [Google Scholar] [CrossRef]
- Wang, B.; Sun, W.; Li, Q.; Li, Y.; Luo, H.; Song, J.; Sun, C.; Qian, J.; Zhu, Y.; Hayward, A.; et al. Genome-wide identification of phenolic acid biosynthetic genes in Salvia miltiorrhiza. Planta 2015, 241, 711–725. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Wan, G.; Liang, Z. Accumulation of salicylic acid-induced phenolic compounds and raised activities of secondary metabolic and antioxidative enzymes in Salvia miltiorrhiza cell culture. J. Biotechnol. 2010, 148, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Dong, J.; Zhang, J.; Dang, X.; Xing, B.; Yang, X. Effects of calcium on synthesis of rosmarinic acid and related enzymes in suspension cultures of Salvia miltiorrhiza. Sheng Wu Gong Cheng Xue Bao 2012, 28, 1359–1369. [Google Scholar] [PubMed]
- Liang, Z.; Ma, Y.; Xu, T.; Cui, B.; Liu, Y.; Guo, Z.; Yang, D. Effects of abscisic acid, gibberellin, ethylene and their interactions on production of phenolic acids in Salvia miltiorrhiza Bunge hairy roots. PLoS ONE 2013, 8, e72806. [Google Scholar] [CrossRef] [PubMed]
- Hao, W.F.; Guo, H.B.; Zhang, J.Y.; Hu, G.G.; Yao, Y.Q.; Dong, J.E. Hydrogen peroxide is involved in salicylic acid-elicited rosmarinic acid production in Salvia miltiorrhiza cell cultures. Sci. World J. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Duan, Y.; Yi, B.; Sun, L.; Lu, B.; Yu, X.; Sun, H.; Zhang, H.; Chen, W. Characterization and expression profiling of cinnamate 4-hydroxylase gene from Salvia miltiorrhiza in rosmarinic acid biosynthesis pathway. Russ. J. Plant Physiol. 2008, 55, 390–399. [Google Scholar] [CrossRef]
- Xiao, Y.; Gao, S.; Di, P.; Chen, J.; Chen, W.; Zhang, L. Methyl jasmonate dramatically enhances the accumulation of phenolic acids in Salvia miltiorrhiza hairy root cultures. Physiol. Plant. 2009, 137, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Yi, B.; Duan, Y.; Sun, L.; Yu, X.; Guo, J.; Chen, W. Characterization and expression profiling of tyrosine aminotransferase gene from Salvia miltiorrhiza (Dan-shen) in rosmarinic acid biosynthesis pathway. Mol. Biol. Rep. 2008, 35, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Yi, B. Molecular Cloning and Regulation Analysis of Important Genes Involved in Tyrosine-branch Pathway of Rosmarinic Acid Metabolism from Salvia miltiorrhza Bunge. Master’s Thesis, Second Military Medical University, Shanghai, China, 2007. [Google Scholar]
- Chen, S.L.; Wang, B.; Sun, W.; Xu, H.B. Genome-Wide Identification and Profiling of Novel Genes Involved in Phenolic Acids Biosynthesis Pathway in Salvia miltiorrhiza, Chinese Academy of Medical Sciences, Institute of Medicinal Plant Development (IMPLAD): Beijing, China, 2013; Unpublished Work.
- Song, J. Function Analysis of the Genes Involved in Rosmarinic Acid Biosynthesis Pathway in Salvia miltiorrhiza Bunge. Ph.D. Thesis, Shaanxi Normal University, Xi’an, China, 2010. [Google Scholar]
- Zhou, Y.J.; Gao, W.; Rong, Q.; Jin, G.; Chu, H.; Liu, W.; Yang, W.; Zhu, Z.; Li, G.; Zhu, G.; et al. Modular pathway engineering of diterpenoid synthases and the mevalonic acid pathway for miltiradiene production. J. Am. Chem. Soc. 2012, 134, 3234–3241. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Liu, Y.; Huang, L.; Zhang, X. Production of miltiradiene by metabolically engineered Saccharomyces cerevisiae. Biotechnol. Bioeng. 2012, 109, 2845–2853. [Google Scholar] [CrossRef] [PubMed]
- Bloch, S.E.; Schmidt-Dannert, C. Construction of a chimeric biosynthetic pathway for the de novo biosynthesis of rosmarinic acid in Escherichia coli. Chembiochem 2014, 15, 2393–2401. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yan, Y.-P.; Wu, Y.-C.; Hua, W.-P.; Chen, C.; Ge, Q.; Wang, Z.-Z. Pathway engineering for phenolic acid accumulations in Salvia miltiorrhiza by combinational genetic manipulation. Metab. Eng. 2014, 21, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds are not available from the authors.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, X.-H.; Ma, Y.; Tang, J.-F.; He, Y.-L.; Liu, Y.-C.; Ma, X.-J.; Shen, Y.; Cui, G.-H.; Lin, H.-X.; Rong, Q.-X.; et al. The Biosynthetic Pathways of Tanshinones and Phenolic Acids in Salvia miltiorrhiza. Molecules 2015, 20, 16235-16254. https://doi.org/10.3390/molecules200916235
Ma X-H, Ma Y, Tang J-F, He Y-L, Liu Y-C, Ma X-J, Shen Y, Cui G-H, Lin H-X, Rong Q-X, et al. The Biosynthetic Pathways of Tanshinones and Phenolic Acids in Salvia miltiorrhiza. Molecules. 2015; 20(9):16235-16254. https://doi.org/10.3390/molecules200916235
Chicago/Turabian StyleMa, Xiao-Hui, Ying Ma, Jin-Fu Tang, Ya-Li He, Yu-Chen Liu, Xiao-Jing Ma, Ye Shen, Guang-Hong Cui, Hui-Xin Lin, Qi-Xian Rong, and et al. 2015. "The Biosynthetic Pathways of Tanshinones and Phenolic Acids in Salvia miltiorrhiza" Molecules 20, no. 9: 16235-16254. https://doi.org/10.3390/molecules200916235





