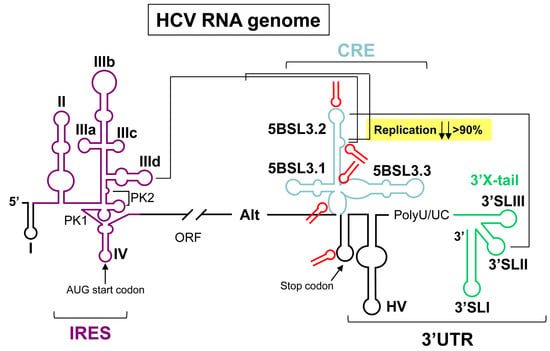
RNA Aptamers as Molecular Tools to Study the Functionality of the Hepatitis C Virus CRE Region
Abstract
:1. Introduction
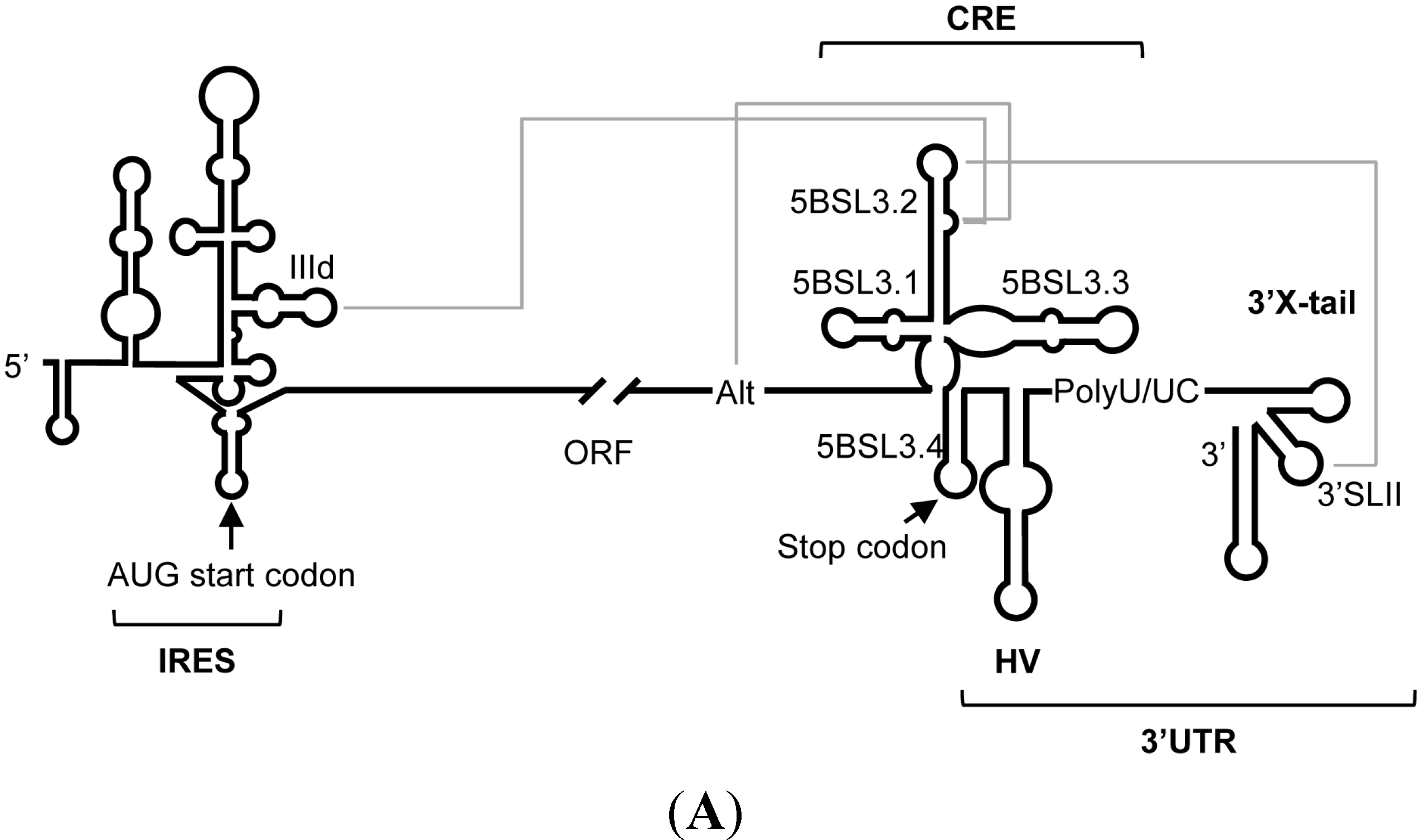

2. Results and Discussion
2.1. Inhibition of HCV Replication by a Collection of RNA Aptamers Targeting the HCV CRE Region
| Aptamer | SEQUENCE (5′→3′) | GROUP | ||||
|---|---|---|---|---|---|---|
| P6-1 | CGUGGACGAGAGCUGGUUGGCGAU | |||||
| P6-2 | GCUGCUGUUACGUACUAAGGUGCGG | |||||
| P6-6 | CGGCUCUGGAUGGCGCUGUUUUGGU | |||||
| P6-8 | CAUUGUGCGACGAGGGCGUGUCCG | |||||
| P6-19 | CGUCCCCGACAGGAUGGGGACAUGG | |||||
| P6-20 | CAACGUGGCGAUGGCUACGAUGUGG | |||||
| P6-23 | CCGCAUCUGCGGACAGGG | |||||
| P6-43 | GCAUCGGAUUGCAGUGCCCU | |||||
| P6-44 | CGCGGCUUCGUUAGCCAUCUGAUG | |||||
| P6-45 | CUGCUGGCUAGUGUCCGG | |||||
| P6-50 | CGGAACGUCUGUUGUGC | |||||
| P6-53 | CAUGUUAGUUACGCGUAGGCGUGCC | |||||
| P6-57 | GCGGCCUGCGAUCUGGAUGCUGCGC | |||||
| P6-64 | CCGAGGGACAGCAGGAGGAGCG | |||||
| P6-76 | GGCAGCUCUAGAGGGUAAUCGGCUCG | |||||
| P6-77 | GUGCUUGCGGUGUUGAGCCCAGCGGU | |||||
| P6-78 | GGUACGGCAUGGCGCUACGAUCGUG | |||||
| P6-79 | GCUAUCCUGGUCCGUGCCG | |||||
| P6-80 | CGCUAGCGUGUUGCAGUAGGCAGAG | |||||
| P6-81 | CAGGAUGAGUACUCCUCGGCGUUGG | |||||
| P6-82 | UAUGCAUUGACGGACGACUGGCCGG | |||||
| P6-83 | UUGGUGACCUUUGUGCUAA | |||||
| P6-84 | CCCUGUGUGGGCUACGUGAG | |||||
| P6-85 | GGGGCGUGUUACGCCUUGUACGAACG | |||||
| P6-86 | GUCGACUGAAGUUUGAAGGA | |||||
| P6-87 | CGUGAUAGUUGUGCUGGCCGAAC | |||||
| P6-88 | CGCUAGAGGUGUUUCUGU | |||||
| P6-89 | CGCCGUGCCAGCUCGGUGCGGCAGG | |||||
| P6-91 | GUCGGCUGUUGACACGUG | |||||
| P6-94 | CGCAGAAUAAGAAUGUGACAG | |||||
| P6-95 | CGCGUGCGCGUUCGUACAG | |||||
| P6-96 | CGUGUUACUGCACAUCGGUG | |||||
| P6-98 | GAUGAGGCCUCGGUAGACAGUGCAG | |||||
| P6-99 | GUGGCCAACGGAACAUGCCGUG | |||||
| P6-100 | CGUGUUACUGCUGGGUACAUCGGUG | |||||
| P6-101 | CGGUAACGUGGCCUUAAGGA | |||||
| P6-102 | CGGCACGAUCUACCGCC | |||||
| P6-103 | GACGUCGUCUGACUCGUGC | |||||
| P7-4 | GGCGUUGGUUUGUAUCGCGGCUUCG | |||||
| P7-15 | CGUCCCAAUUGACACGUGGCAGCCG | |||||
| P7-49 | CCGUGCGUGCGUCACGGCCUGG | |||||
| P7-90 | CGGCCGUUGCUGGAGUGCCGC | |||||
| P7-93 | CAACUGCUCGUGAGAGUGGCU | |||||
| P7-97 | GACUCUAUUGAGGGACGAUGG | |||||

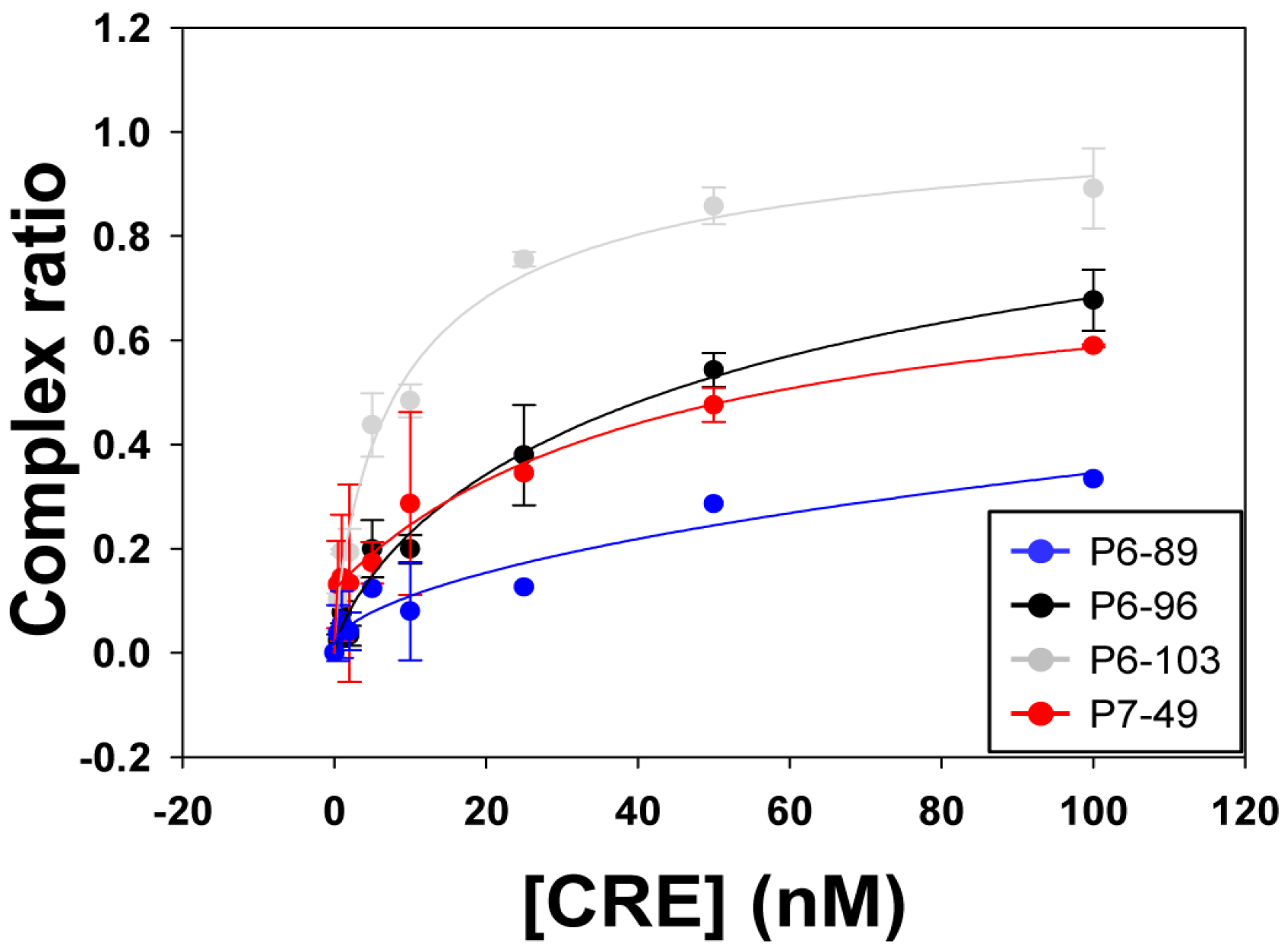
2.2. Biochemical Analysis of the Selected Aptamers
| Aptamer | Kd (nM) ± SD | Bmax ± SD |
|---|---|---|
| P6-89 | 1706.34 ± 230.15 | n.d. |
| P6-96 | 62.67 ± 0.74 | 1.15 ± 0.05 |
| P6-103 | 9.47 ± 3.49 | 1.04 ± 0.14 |
| P7-49 | 43.63 ± 16.22 | 1.07 ± 0.07 |

2.3. Aptamers Targeting the 5BSL3.2 Domain Compete with the Recruitment of the Viral RNA-Dependent RNA Polymerase by the CRE Region
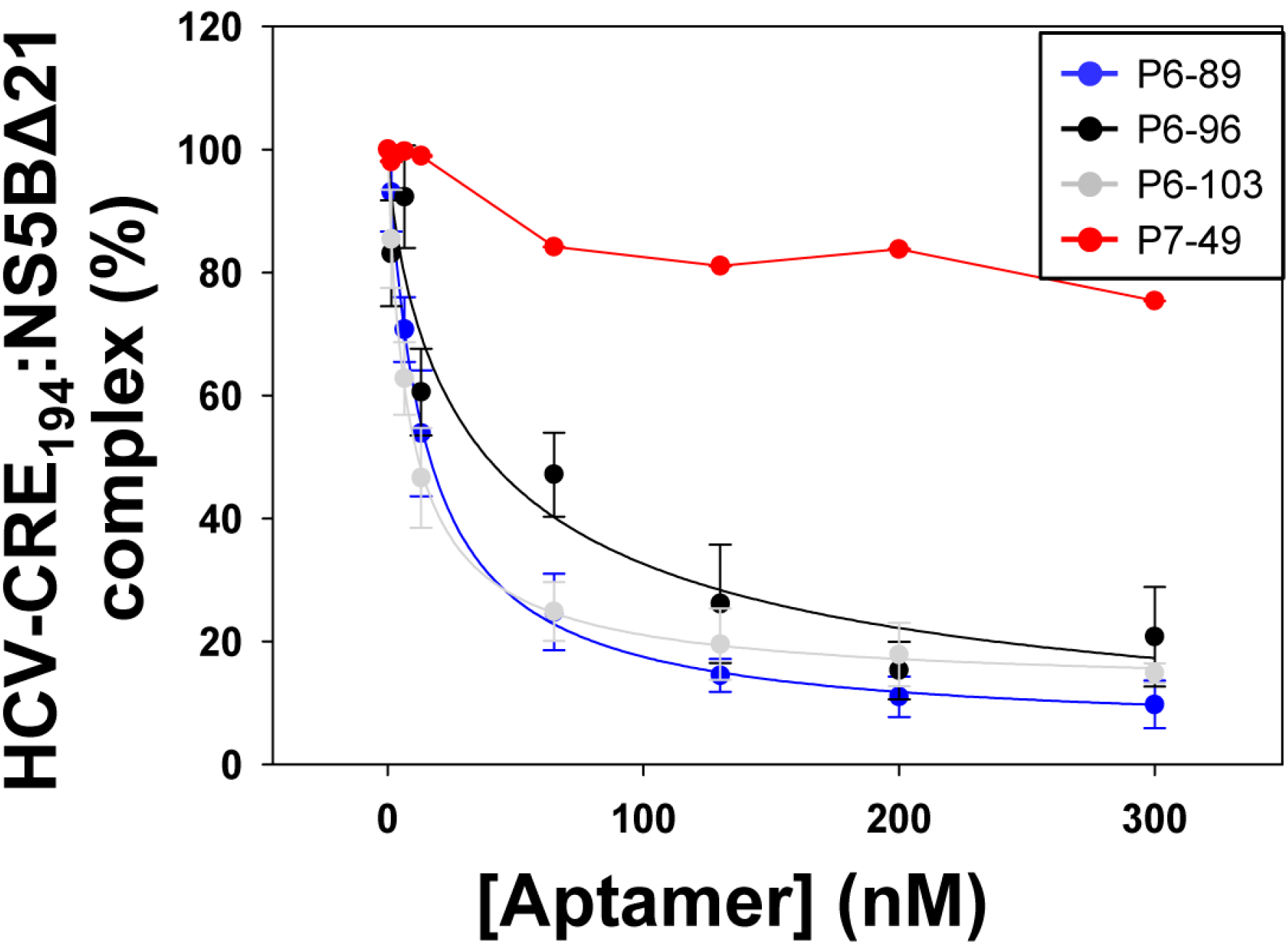
| Aptamer | EC50 (nM) a | HCV-CRE194:NS5BΔ21 Complex (%) b |
|---|---|---|
| P6-89 | 14.59 ± 1.11 | 5.09 ± 2.04 |
| P6-96 | 38.68 ± 5.63 | 0.00 ± 4.15 |
| P6-103 | 8.57 ± 0.58 | 11.98 ± 1.46 |
| P7-49 | n.d. | 75.37 ± 3.05 |
2.4. Effect of the Aptamers on HCV Translation

2.5. Discussion

3. Experimental Section
3.1. Cell Lines and HCV Constructs
3.2. DNA Templates and RNA Synthesis
3.3. Cell Transfection
3.4. Quantification of the HCV RNA Replicon
3.5. Binding Assays
3.6. Aptamer Competition with the Binding of the NS5BΔ21 to the HCV-CRE194
3.7. Luciferase Assays
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Santosh, B.; Yadava, P.K. Nucleic acid aptamers: Research tools in disease diagnostics and therapeutics. Biomed. Res. Int. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Binning, J.M.; Leung, D.W.; Amarasinghe, G.K. Aptamers in virology: Recent advances and challenges. Front. Microbiol. 2012, 3, 29. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Kim, J.H.; Lee, S.W. Prospects for nucleic acid-based therapeutics against hepatitis C virus. World J. Gastroenterol. 2013, 19, 8949–8962. [Google Scholar] [CrossRef] [PubMed]
- Kieffer, T.L.; Sarrazin, C.; Miller, J.S.; Welker, M.W.; Forestier, N.; Reesink, H.W.; Kwong, A.D.; Zeuzem, S. Telaprevir and pegylated interferon-alpha-2a inhibit wild-type and resistant genotype 1 hepatitis C virus replication in patients. Hepatology 2007, 46, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Kieffer, T.L.; Kwong, A.D.; Picchio, G.R. Viral resistance to specifically targeted antiviral therapies for hepatitis C (STAT-CS). J. Antimicrob. Chemother. 2010, 65, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Choo, Q.L.; Kuo, G.; Weiner, A.J.; Overby, L.R.; Bradley, D.W.; Houghton, M. Isolation of a cdna clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 1989, 244, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Takamizawa, A.; Mori, C.; Fuke, I.; Manabe, S.; Murakami, S.; Fujita, J.; Onishi, E.; Andoh, T.; Yoshida, I.; Okayama, H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J. Virol. 1991, 65, 1105–1113. [Google Scholar] [PubMed]
- Tsukiyama-Kohara, K.; Iizuka, N.; Kohara, M.; Nomoto, A. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 1992, 66, 1476–1483. [Google Scholar] [PubMed]
- Wang, C.; Sarnow, P.; Siddiqui, A. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J. Virol. 1993, 67, 3338–3344. [Google Scholar] [PubMed]
- Wang, C.; Sarnow, P.; Siddiqui, A. A conserved helical element is essential for internal initiation of translation of hepatitis C virus RNA. J. Virol. 1994, 68, 7301–7307. [Google Scholar] [PubMed]
- Wang, C.; Le, S.Y.; Ali, N.; Siddiqui, A. An rna pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA 1995, 1, 526–537. [Google Scholar] [PubMed]
- Friebe, P.; Lohmann, V.; Krieger, N.; Bartenschlager, R. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 2001, 75, 12047–12057. [Google Scholar] [CrossRef] [PubMed]
- Friebe, P.; Bartenschlager, R. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 2002, 76, 5326–5338. [Google Scholar] [CrossRef] [PubMed]
- Yi, M.; Lemon, S.M. 3′ nontranslated RNA signals required for replication of hepatitis C virus RNA. J. Virol. 2003, 77, 3557–3568. [Google Scholar] [CrossRef] [PubMed]
- Tuplin, A.; Wood, J.; Evans, D.J.; Patel, A.H.; Simmonds, P. Thermodynamic and phylogenetic prediction of RNA secondary structures in the coding region of hepatitis C virus. RNA 2002, 8, 824–841. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Shin, H.; Wimmer, E.; Paul, A.V. Cis-acting RNA signals in the NS5B c-terminal coding sequence of the hepatitis C virus genome. J. Virol. 2004, 78, 10865–10877. [Google Scholar] [CrossRef] [PubMed]
- You, S.; Stump, D.D.; Branch, A.D.; Rice, C.M. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J. Virol. 2004, 78, 1352–1366. [Google Scholar] [CrossRef] [PubMed]
- You, S.; Rice, C.M. 3′ RNA elements in hepatitis C virus replication: Kissing partners and long poly(U). J. Virol. 2008, 82, 184–195. [Google Scholar] [CrossRef] [PubMed]
- Friebe, P.; Boudet, J.; Simorre, J.P.; Bartenschlager, R. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 2005, 79, 380–392. [Google Scholar] [CrossRef] [PubMed]
- Romero-López, C.; Berzal-Herranz, A. The functional RNA domain 5BSL3.2 within the NS5B coding sequence influences hepatitis C virus IRES-mediated translation. Cell. Mol. Life Sci. 2012, 69, 103–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero-López, C.; Berzal-Herranz, A. A long-range RNA-RNA interaction between the 5′ and 3′ ends of the HCV genome. RNA 2009, 15, 1740–1752. [Google Scholar] [CrossRef] [PubMed]
- Diviney, S.; Tuplin, A.; Struthers, M.; Armstrong, V.; Elliott, R.M.; Simmonds, P.; Evans, D.J. A hepatitis C virus cis-acting replication element forms a long-range RNA-RNA interaction with upstream RNA sequences in NS5B. J. Virol. 2008, 82, 9008–9022. [Google Scholar] [CrossRef] [PubMed]
- Shetty, S.; Stefanovic, S.; Mihailescu, M.R. Hepatitis C virus RNA: Molecular switches mediated by long-range RNA-RNA interactions? Nucleic Acids Res. 2013, 41, 2526–2540. [Google Scholar] [CrossRef] [PubMed]
- Romero-López, C.; Barroso-delJesus, A.; García-Sacristán, A.; Briones, C.; Berzal-Herranz, A. The folding of the hepatitis C virus internal ribosome entry site depends on the 3′-end of the viral genome. Nucleic Acids Res. 2012, 40, 11697–11713. [Google Scholar] [CrossRef] [PubMed]
- Tuplin, A.; Struthers, M.; Simmonds, P.; Evans, D.J. A twist in the tail: SHAPE mapping of long-range interactions and structural rearrangements of RNA elements involved in HCV replication. Nucleic Acids Res. 2012, 40, 6908–6921. [Google Scholar] [CrossRef] [PubMed]
- Romero-López, C.; Barroso-delJesus, A.; García-Sacristán, A.; Briones, C.; Berzal-Herranz, A. End-to-end crosstalk within the hepatitis C virus genome mediates the conformational switch of the 3′X-tail region. Nucleic Acids Res. 2014, 42, 567–582. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yamada, O.; Sakamoto, T.; Yoshida, H.; Araki, H.; Murata, T.; Shimotohno, K. Inhibition of hepatitis C virus replication by Pol III-directed overexpression of RNA decoys corresponding to stem-loop structures in the NS5B coding region. Virology 2005, 342, 276–285. [Google Scholar] [CrossRef] [PubMed]
- Kanamori, H.; Yuhashi, K.; Ohnishi, S.; Koike, K.; Kodama, T. RNA-dependent RNA polymerase of hepatitis C virus binds to its coding region RNA stem-loop structure, 5BSL3.2, and its negative strand. J. Gen. Virol. 2010, 91, 1207–1212. [Google Scholar] [CrossRef] [PubMed]
- Oakland, T.E.; Haselton, K.J.; Randall, G. Ewsr1 binds the hepatitis C virus cis-acting replication element and is required for efficient viral replication. J. Virol. 2013, 87, 6625–6634. [Google Scholar] [CrossRef] [PubMed]
- Marton, S.; Reyes-Darias, J.A.; Sánchez-Luque, F.J.; Romero-López, C.; Berzal-Herranz, A. In vitro and ex vivo selection procedures for identifying potentially therapeutic DNA and RNA molecules. Molecules 2010, 15, 4610–4638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marton, S.; Romero-López, C.; Berzal-Herranz, A. RNA aptamer-mediated interference of HCV replication by targeting the CRE-5BSL3.2 domain. J. Viral Hepat. 2013, 20, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Marton, S.; Berzal-Herranz, B.; Garmendia, E.; Cueto, F.J.; Berzal-Herranz, A. Anti-HCV RNA aptamers targeting the genomic cis-acting replication element. Pharmaceuticals (Basel) 2011, 5, 49–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lohmann, V.; Korner, F.; Koch, J.; Herian, U.; Theilmann, L.; Bartenschlager, R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 1999, 285, 110–113. [Google Scholar] [CrossRef] [PubMed]
- Larrea, E.; Aldabe, R.; Molano, E.; Fernandez-Rodriguez, C.M.; Ametzazurra, A.; Civeira, M.P.; Prieto, J. Altered expression and activation of signal transducers and activators of transcription (STATS) in hepatitis C virus infection: In vivo and in vitro studies. Gut 2006, 55, 1188–1196. [Google Scholar] [CrossRef] [PubMed]
- Romero-López, C.; Barroso-delJesus, A.; Puerta-Fernández, E.; Berzal-Herranz, A. Interfering with hepatitis C virus IRES activity using RNA molecules identified by a novel in vitro selection method. Biol. Chem. 2005, 386, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Romero-López, C.; Díaz-González, R.; Barroso-delJesus, A.; Berzal-Herranz, A. Inhibition of HCV replication and IRES-dependent translation by an RNA molecule. J. Gen. Virol. 2009, 90, 1659–1669. [Google Scholar] [CrossRef] [PubMed]
- Harmanci, A.O.; Sharma, G.; Mathews, D.H. Turbofold: Iterative probabilistic estimation of secondary structures for multiple RNA sequences. BMC Bioinform. 2011, 12. [Google Scholar] [CrossRef] [PubMed]
- Brunel, C.; Marquet, R.; Romby, P.; Ehresmann, C. RNA loop-loop interactions as dynamic functional motifs. Biochimie 2002, 84, 925–944. [Google Scholar] [CrossRef]
- Lohmann, V.; Hoffmann, S.; Herian, U.; Penin, F.; Bartenschlager, R. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 2003, 77, 3007–3019. [Google Scholar] [CrossRef] [PubMed]
- Kean, K.M. The role of mRNA 5′-noncoding and 3′-end sequences on 40S ribosomal subunit recruitment, and how RNA viruses successfully compete with cellular mRNAs to ensure their own protein synthesis. Biol. Cell 2003, 95, 129–139. [Google Scholar] [CrossRef]
- Darfeuille, F.; Reigadas, S.; Hansen, J.B.; Orum, H.; Di Primo, C.; Toulme, J.J. Aptamers targeted to an RNA hairpin show improved specificity compared to that of complementary oligonucleotides. Biochemistry 2006, 45, 12076–12082. [Google Scholar] [CrossRef] [PubMed]
- Blight, K.J.; McKeating, J.A.; Rice, C.M. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 2002, 76, 13001–13014. [Google Scholar] [CrossRef] [PubMed]
- Romero-López, C.; Díaz-González, R.; Berzal-Herranz, A. Inhibition of hepatitis C virus internal ribosome entry site-mediated translation by an RNA targeting the conserved IIIf domain. Cell. Mol. Life Sci. 2007, 64, 2994–3006. [Google Scholar] [CrossRef] [PubMed]
- Van den Hoff, M.J.; Moorman, A.F.; Lamers, W.H. Electroporation in “intracellular” buffer increases cell survival. Nucleic Acids Res. 1992, 20, 2902. [Google Scholar] [CrossRef] [PubMed]
- Martell, M.; Gomez, J.; Esteban, J.I.; Sauleda, S.; Quer, J.; Cabot, B.; Esteban, R.; Guardia, J. High-throughput real-time reverse transcription-pcr quantitation of hepatitis C virus RNA. J. Clin. Microbiol. 1999, 37, 327–332. [Google Scholar] [PubMed]
- Barroso-delJesus, A.; Romero-López, C.; Lucena-Aguilar, G.; Melen, G.J.; Sanchez, L.; Ligero, G.; Berzal-Herranz, A.; Menendez, P. Embryonic stem cell-specific mir302–367 cluster: Human gene structure and functional characterization of its core promoter. Mol. Cell. Biol. 2008, 28, 6609–6619. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples corresponding to plasmid constructs are available from the authors.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández-Sanlés, A.; Berzal-Herranz, B.; González-Matamala, R.; Ríos-Marco, P.; Romero-López, C.; Berzal-Herranz, A. RNA Aptamers as Molecular Tools to Study the Functionality of the Hepatitis C Virus CRE Region. Molecules 2015, 20, 16030-16047. https://doi.org/10.3390/molecules200916030
Fernández-Sanlés A, Berzal-Herranz B, González-Matamala R, Ríos-Marco P, Romero-López C, Berzal-Herranz A. RNA Aptamers as Molecular Tools to Study the Functionality of the Hepatitis C Virus CRE Region. Molecules. 2015; 20(9):16030-16047. https://doi.org/10.3390/molecules200916030
Chicago/Turabian StyleFernández-Sanlés, Alba, Beatriz Berzal-Herranz, Rodrigo González-Matamala, Pablo Ríos-Marco, Cristina Romero-López, and Alfredo Berzal-Herranz. 2015. "RNA Aptamers as Molecular Tools to Study the Functionality of the Hepatitis C Virus CRE Region" Molecules 20, no. 9: 16030-16047. https://doi.org/10.3390/molecules200916030