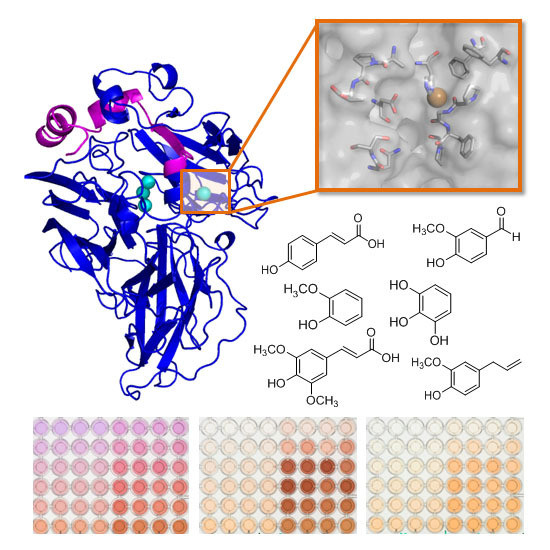
Exploring the Oxidation of Lignin-Derived Phenols by a Library of Laccase Mutants
Abstract
:1. Introduction
2. Results and Discussion
| Code | Name | Structure | Wavelength (nm) | Time (h) |
|---|---|---|---|---|
| Phenols | ||||
| 1 | Phenol | 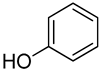 | 420 | 24 |
| 2 | Catechol | 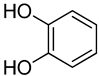 | 400 | 2 |
| 3 | Guaiacol | 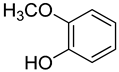 | 470 | 2 |
| 4 | Pyrogallol | 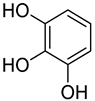 | 450 | 2 |
| 5 | 2,6-Dimethoxyphenol | 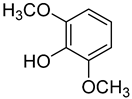 | 470 | 0.25 a |
| p-Hydroxybenzoic Acids | ||||
| 6 | 4-Hydroxybenzoic acid |  | 260 | 24 |
| 7 | 3,4-Dihydroxybenzoic acid | 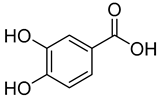 | 410 | 2 |
| 8 | Vanillic acid |  | 350 | 4 |
| 9 | Gallic acid |  | 450 | 2 |
| 10 | Syringic acid |  | 360 | 2 |
| p-Hydroxycinnamic Acids | ||||
| 11 | p-Coumaric acid | 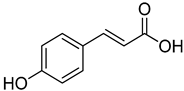 | 260 | 24 |
| 12 | Caffeic acid | 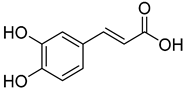 | 390 | 2 |
| 13 | Ferulic acid | 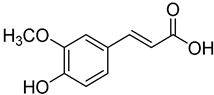 | 430 | 2 |
| 14 | Sinapic acid | 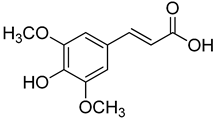 | 520 | 2 |
| p-Hydroxyacetophenones | ||||
| 15 | 4-hydroxyacetophenone | 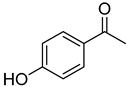 | 260 | 24 |
| 16 | Acetovanillone | 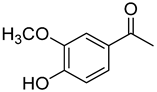 | 300 | 24 |
| 17 | Acetosyringone |  | 520 | 8 |
| p-Hydroxybenzaldehydes | ||||
| 18 | 4-hydroxybenzaldehyde | 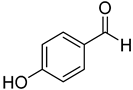 | 260 | 24 |
| 19 | Vanillin | 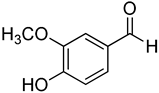 | 410 | 24 |
| 20 | Syringaldehyde | 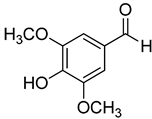 | 370 | 4 |
| Methyl p-Hydroxybenzoates | ||||
| 21 | Methyl vanillate | 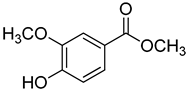 | 370 | 24 |
| 22 | Methyl syringate | 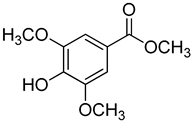 | 390 | 1 |
| Methyl p-Hydroxycinnamates | ||||
| 23 | Methyl p-coumarate |  | 400 | 24 |
| 24 | Methyl ferulate | 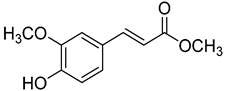 | 370 | 1 |
| 25 | Methyl sinapate |  | 410 | 4 |
| Others | ||||
| 26 | Eugenol | 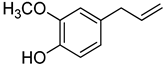 | 370 | 1 |
| 27 | Resveratrol | 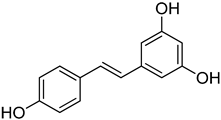 | 400 | 1 |
| 28 | Quercetin |  | 290 | 4 |
2.1. Production of Laccase Mutants
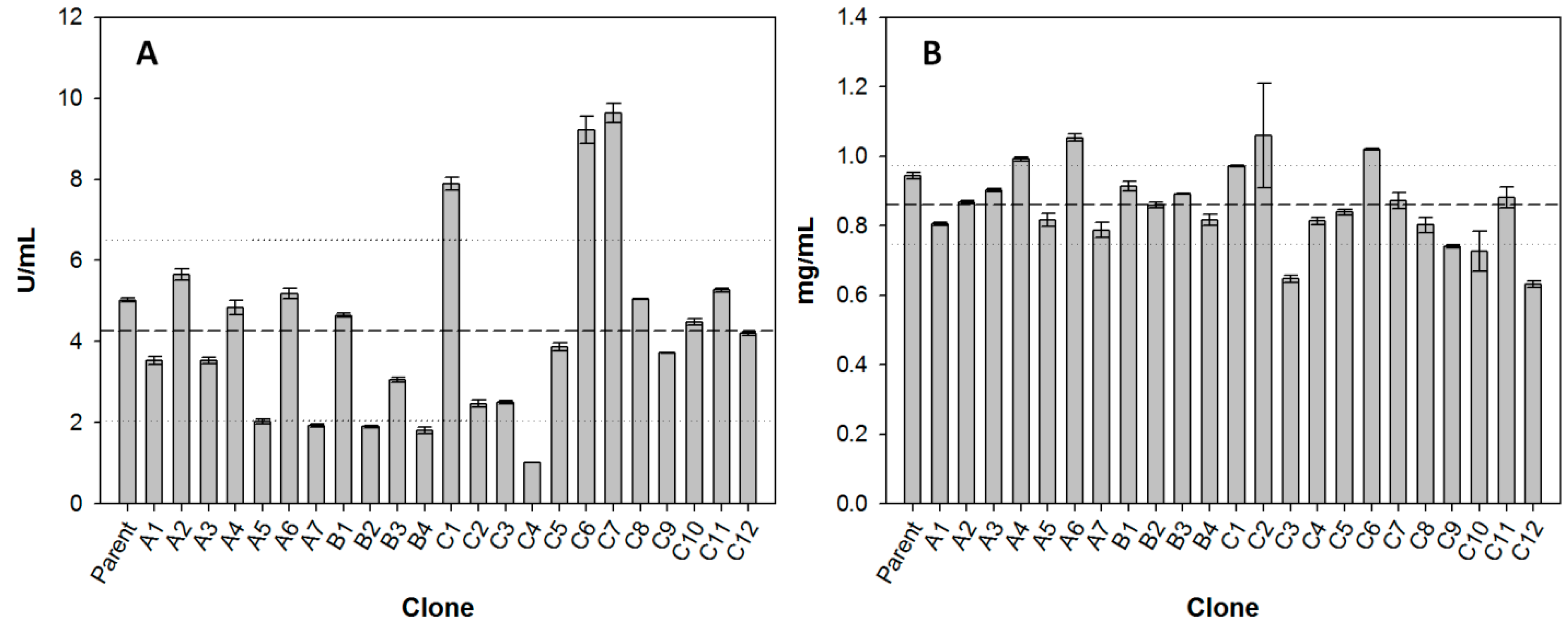
2.2. Multi-Screening with Phenolic Compounds
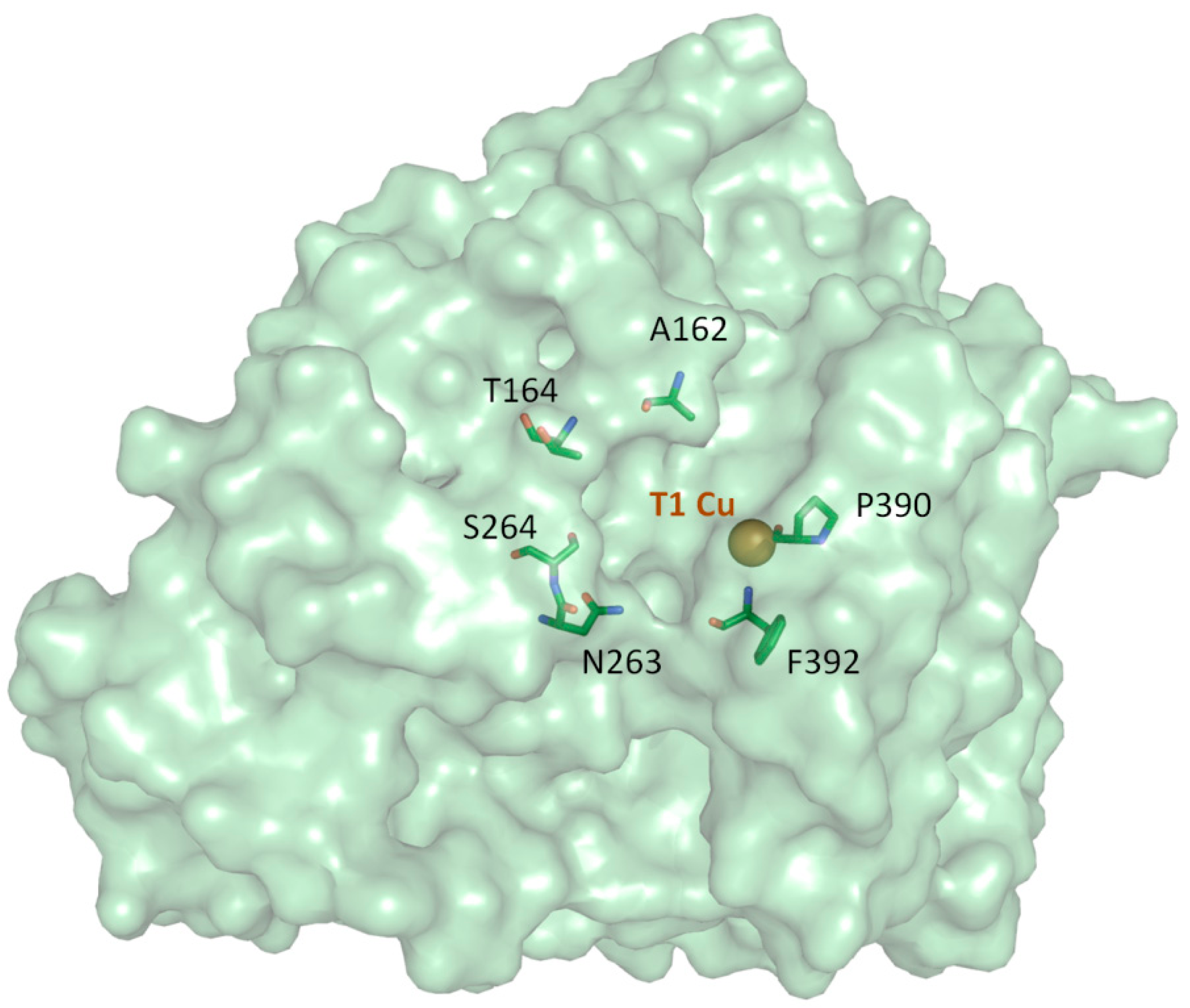
2.3. Comparison of Laccase Mutants
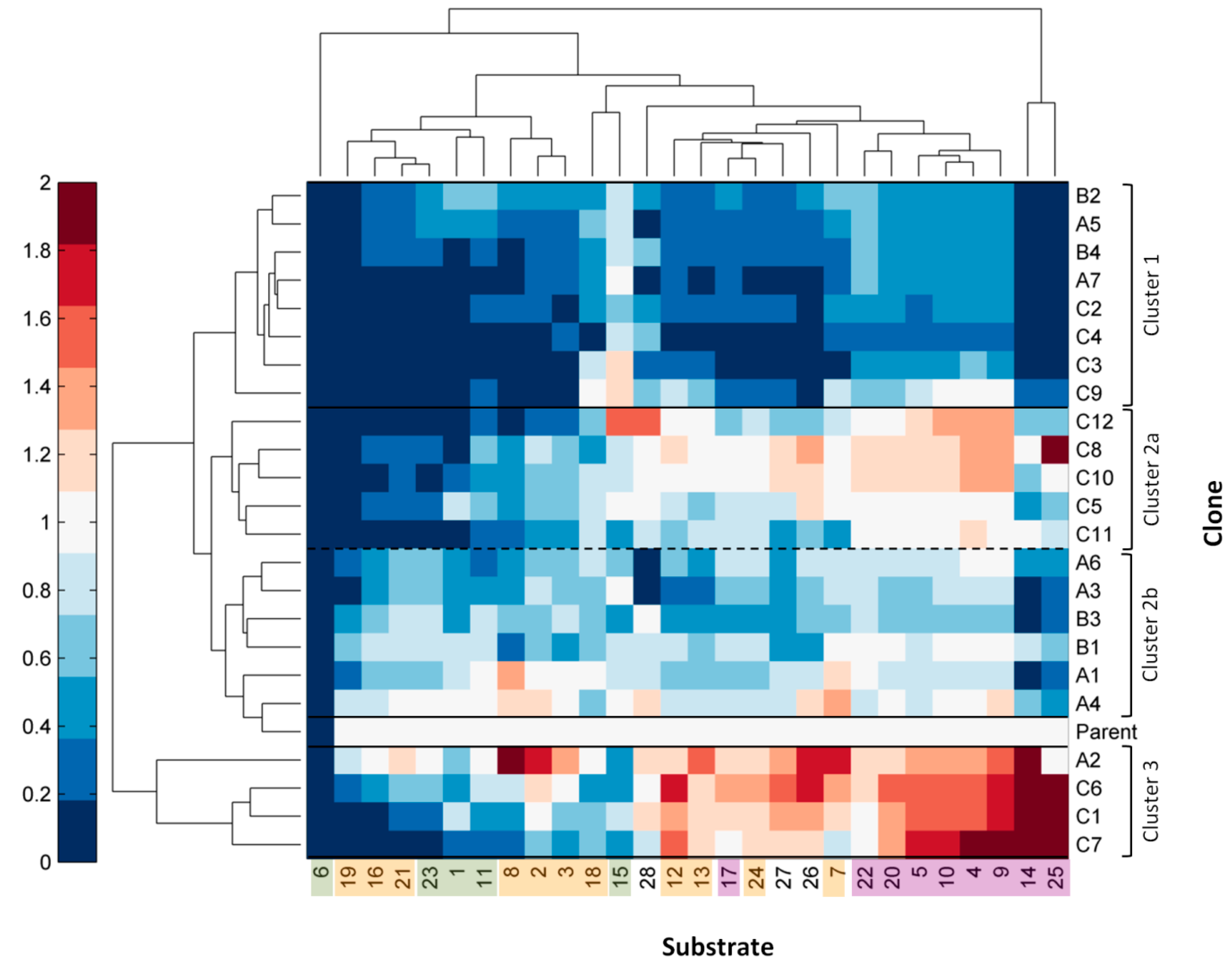
3. Experimental Section
3.1. Construction and Screening of Mutant Libraries
3.2. Production of Laccase Mutants
3.3. Multi-Screening with Phenolic Compounds
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Giardina, P.; Faraco, V.; Pezzella, C.; Piscitelli, A.; Vanhulle, S.; Sannia, G. Laccases: A never-ending story. Cell. Mol. Life Sci. 2010, 67, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Camarero, S.; Martínez, M.J.; Martínez, A.T. Understanding lignin biodegradation for the improved utilization of plant biomass in modern biorefineries. Biofuels Bioprod. Biorefin. 2014, 8, 615–625. [Google Scholar] [CrossRef]
- Jurado, M.; Prieto, A.; Martínez-Alcalá, A.; Martinez, A.T.; Martínez, M.J. Laccase detoxification of steam-exploded wheat straw for second generation bioethanol. Bioresour. Technol. 2009, 100, 6378–6384. [Google Scholar] [CrossRef] [PubMed]
- Ibarra, D.; Camarero, S.; Romero, J.; Martínez, M.J.; Martínez, A.T. Integrating laccase-mediator treatment into an industrial-type sequence for totally chlorine-free bleaching of eucalypt kraft pulp. J. Chem. Technol. Biotechnol. 2006, 81, 1159–1165. [Google Scholar] [CrossRef]
- Fillat, U.; Prieto, A.; Camarero, S.; Martinez, A.T.; Martínez, M.J. Biodeinking of flexographic inks by fungal laccases using synthetic and natural mediators. Biochem. Eng. J. 2012, 67, 97–103. [Google Scholar] [CrossRef]
- Aracri, E.; Fillat, A.; Colom, J.F.; Gutiérrez, A.; del Río, J.C.; Martinez, A.T.; Vidal, T. Enzymatic grafting of simple phenols on flax and sisal pulp fibres using laccases. Bioresour. Technol. 2010, 101, 8211–8216. [Google Scholar] [CrossRef] [PubMed]
- Božič, M.; Gorgieva, S.; Kokol, V. Laccase-mediated functionalization of chitosan by caffeic and gallic acids for modulating antioxidant and antimicrobial properties. Carbohydr. Polym. 2012, 87, 2388–2398. [Google Scholar] [CrossRef]
- Kudanga, T.; Prasetyo, E.N.; Sipilä, J.; Nousiainen, P.; Widsten, P.; Kandelbauer, A.; Nyanhongo, G.S.; Guebitz, G. Laccase-Mediated Wood Surface Functionalization. Eng. Life Sci. 2008, 8, 297–302. [Google Scholar] [CrossRef]
- Riva, S. Laccases: Blue enzymes for green chemistry. Trends Biotechnol. 2006, 24, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Kunamneni, A.; Camarero, S.; García-Burgos, C.; Plou, F.J.; Ballesteros, A.; Alcalde, M. Engineering and Applications of fungal laccases for organic synthesis. Microb. Cell Fact. 2008, 7, 32. [Google Scholar] [CrossRef] [PubMed]
- Polak, J.; Jarosz-Wilkolazka, A. Fungal laccases as green catalysts for dye synthesis. Process Biochem. 2012, 47, 1295–1307. [Google Scholar] [CrossRef]
- Mogharabi, M.; Faramarzi, M.A. Laccase and Laccase-Mediated Systems in the Synthesis of Organic Compounds. Adv. Synth. Catal. 2014, 356, 897–927. [Google Scholar] [CrossRef]
- Witayakran, S.; Ragauskas, A.J. One-pot synthesis of 1,4-naphthoquinones and related structures with laccase. Green Chem. 2007, 9, 475–480. [Google Scholar] [CrossRef]
- Witayakran, S.; Zettili, A.; Ragauskas, A.J. Laccase-generated quinones in naphthoquinone synthesis via Diels-Alder reaction. Tetrahedron Lett. 2007, 48, 2983–2987. [Google Scholar] [CrossRef]
- Camarero, S.; Ibarra, D.; Martínez, M.J.; Martinez, A.T. Lignin-Derived Compounds as Efficient Laccase Mediators for Decolorization of Different Types of Recalcitrant Dyes. Appl. Environ. Microbiol. 2005, 71, 1775–1784. [Google Scholar] [CrossRef] [PubMed]
- Camarero, S.; Ibarra, D.; Martinez, A.T.; Romero, J.; Gutiérrez, A.; del Río, J.C. Paper pulp delignification using laccase and natural mediators. Enzyme Microb. Technol. 2007, 40, 1264–1271. [Google Scholar] [CrossRef] [Green Version]
- Camarero, S.; Cañas, A.I.; Nousiainen, P.; Record, E.; Lomascolo, A.; Martínez, M.J.; Martinez, A.T. p-Hydroxycinnamic acids as natural mediators for laccase oxidation of recalcitrant compounds. Environ. Sci. Technol. 2008, 42, 6703–6709. [Google Scholar] [CrossRef] [PubMed]
- Cañas, A.I.; Camarero, S. Laccases and their natural mediators: Biotechnological tools for sustainable eco-friendly processes. Biotechnol. Adv. 2010, 28, 694–705. [Google Scholar] [CrossRef] [PubMed]
- Fillat, A.; Colom, J.F.; Vidal, T. A new approach to the biobleaching of flax pulp with laccase using natural mediators. Bioresour. Technol. 2010, 101, 4104–4110. [Google Scholar] [CrossRef] [PubMed]
- Babot, E.D.; Rico, A.; Rencoret, J.; Kalum, L.; Lund, H.; Romero, J.; del Río, J.C.; Martinez, A.T.; Gutiérrez, A. Towards industrially-feasible delignification and pitch removal by treating paper pulp with Myceliophthora thermophila laccase and a phenolic mediator. Bioresour. Technol. 2011, 102, 6717–6722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosado, T.; Bernardo, P.; Koci, K.; Coelho, A.V; Robalo, M.P.; Martins, L.O. Methyl syringate: An efficient phenolic mediator for bacterial and fungal laccases. Bioresour. Technol. 2012, 124, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Pardo, I.; Camarero, S. Laccase engineering by rational and evolutionary design. Cell. Mol. Life Sci. 2015, 72, 897–910. [Google Scholar] [CrossRef] [PubMed]
- Maté, D.; García-Burgos, C.; García-Ruiz, E.; Ballesteros, A.O.; Camarero, S.; Alcalde, M. Laboratory Evolution of High-Redox Potential Laccases. Chem. Biol. 2010, 17, 1030–1041. [Google Scholar] [CrossRef] [PubMed]
- Camarero, S.; Pardo, I.; Cañas, A.I.; Molina, P.; Record, E.; Martinez, A.T.; Martinez, M.J.; Alcalde, M. Engineering Platforms for Directed Evolution of Laccase from Pycnoporus cinnabarinus. Appl. Environ. Microbiol. 2012, 78, 1370–1384. [Google Scholar] [CrossRef] [PubMed]
- Pardo, I.; Vicente, A.I.; Mate, D.M.; Alcalde, M.; Camarero, S. Development of chimeric laccases by directed evolution. Biotechnol. Bioeng. 2012, 109, 2978–2986. [Google Scholar] [CrossRef] [PubMed]
- Mathew, S.; Abraham, T.E. Bioconversions of Ferulic Acid, an Hydroxycinnamic Acid. Crit. Rev. Microbiol. 2006, 32, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Niwa, T.; Doi, U.; Osawa, T. Formation of thomasidioic acid from dehydrosinapinic acid dilactone under neutral conditions, and a remaining inhibitory activity against peroxynitrite-mediated protein nitration. Bioorg. Med. Chem. Lett. 2002, 12, 963–965. [Google Scholar] [CrossRef]
- Yamanaka, D.; Tamiya, Y.; Motoi, M.; Ishibashi, K.; Miura, N.N.; Adachi, Y.; Ohno, N. The effect of enzymatically polymerised polyphenols on CD4 binding and cytokine production in murine splenocytes. PLoS ONE 2012, 7, e36025. [Google Scholar] [CrossRef] [PubMed]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Pardo, I.; Chanagá, X.; Vicente, A.I.; Alcalde, M.; Camarero, S. New colorimetric screening assays for the directed evolution of fungal laccases to improve the conversion of plant biomass. BMC Biotechnol. 2013, 13, 90. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Dannert, C.; Arnold, F.H. Directed evolution of industrial enzymes. Trends Biotechnol. 1999, 17, 135–136. [Google Scholar] [CrossRef]
- García-Ruiz, E.; Maté, D.; Ballesteros, A.; Martinez, A.T.; Alcalde, M. Evolving thermostability in mutant libraries of ligninolytic oxidoreductases expressed in yeast. Microb. Cell Fact. 2010, 9, 17. [Google Scholar] [CrossRef] [PubMed]
- Xu, F. Oxidation of phenols, anilines, and benzenethiols by fungal laccases: Correlation between activity and redox potentials as well as halide inhibition. Biochemistry 1996, 35, 7608–7614. [Google Scholar] [CrossRef] [PubMed]
- D’Annibale, A.; Celletti, D.; Felici, M.; Di Mattia, E.; Giovannozzi-Sermanni, G. Substrate specificity of laccase from Lentinus edodes. Acta Biotechnol. 1996, 16, 257–270. [Google Scholar] [CrossRef]
- Edens, W.; Goins, T.Q.; Dooley, D.; Henson, M. Purification and characterization of a secreted laccase of Gaeumannomyces graminis var. tritici. Appl. Environ. Microbiol. 1999, 65, 3071–3074. [Google Scholar] [PubMed]
- Tadesse, M.A.; D’Annibale, A.; Galli, C.; Gentili, P.; Sergi, F. An assessment of the relative contributions of redox and steric issues to laccase specificity towards putative substrates. Org. Biomol. Chem. 2008, 6, 868–878. [Google Scholar] [CrossRef] [PubMed]
- Monza, E.; Lucas, M.F.; Camarero, S.; Alejaldre, L.C.; Martinez, A.T.; Guallar, V. Insights into laccase engineering from molecular simulations: Toward a binding-focused strategy. J. Phys. Chem. Lett. 2015, 6, 1447–1453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camarero, S.; Galletti, G.C.; Martinez, A.T. Demonstration of In Situ Oxidative Degradation of Lignin Side Chains by Two White-rot Fungi Using Analytical Pyrolysis of Methylated Wheat Straw. Rapid Commun. Mass Spectrom. 1997, 11, 331–334. [Google Scholar] [CrossRef]
- Bertrand, T.; Jolivalt, C.; Briozzo, P.; Caminade, E.; Joly, N.; Madzak, C.; Mougin, C. Crystal Structure of a Four-Copper Laccase Complexed with an Arylamine: Insights into Substrate Recognition and Correlation with Kinetics. Biochemistry 2002, 41, 7325–7333. [Google Scholar] [CrossRef] [PubMed]
- Galli, C.; Gentili, P.; Jolivalt, C.; Madzak, C.; Vadalà, R. How is the reactivity of laccase affected by single-point mutations? Engineering laccase for improved activity towards sterically demanding substrates. Appl. Microbiol. Biotechnol. 2011, 91, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Alcalde, M. Mutagenesis Protocols in Saccharomyces cerevisiae by in Vivo Overlap Extension. Methods Mol. Biol. 2010, 634, 3–14. [Google Scholar] [PubMed]
- Sample Availability: Samples are not available from the authors.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pardo, I.; Camarero, S. Exploring the Oxidation of Lignin-Derived Phenols by a Library of Laccase Mutants. Molecules 2015, 20, 15929-15943. https://doi.org/10.3390/molecules200915929
Pardo I, Camarero S. Exploring the Oxidation of Lignin-Derived Phenols by a Library of Laccase Mutants. Molecules. 2015; 20(9):15929-15943. https://doi.org/10.3390/molecules200915929
Chicago/Turabian StylePardo, Isabel, and Susana Camarero. 2015. "Exploring the Oxidation of Lignin-Derived Phenols by a Library of Laccase Mutants" Molecules 20, no. 9: 15929-15943. https://doi.org/10.3390/molecules200915929