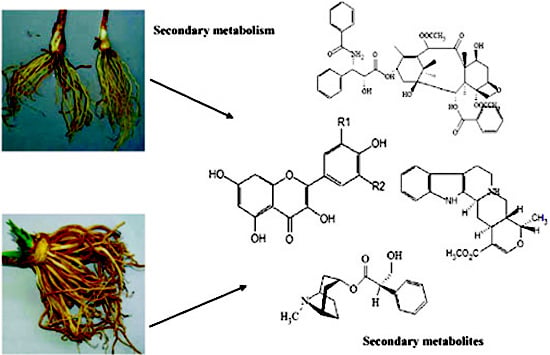
Tilting Plant Metabolism for Improved Metabolite Biosynthesis and Enhanced Human Benefit
Abstract
:1. Introduction
2. Secondary Metabolism and Metabolites
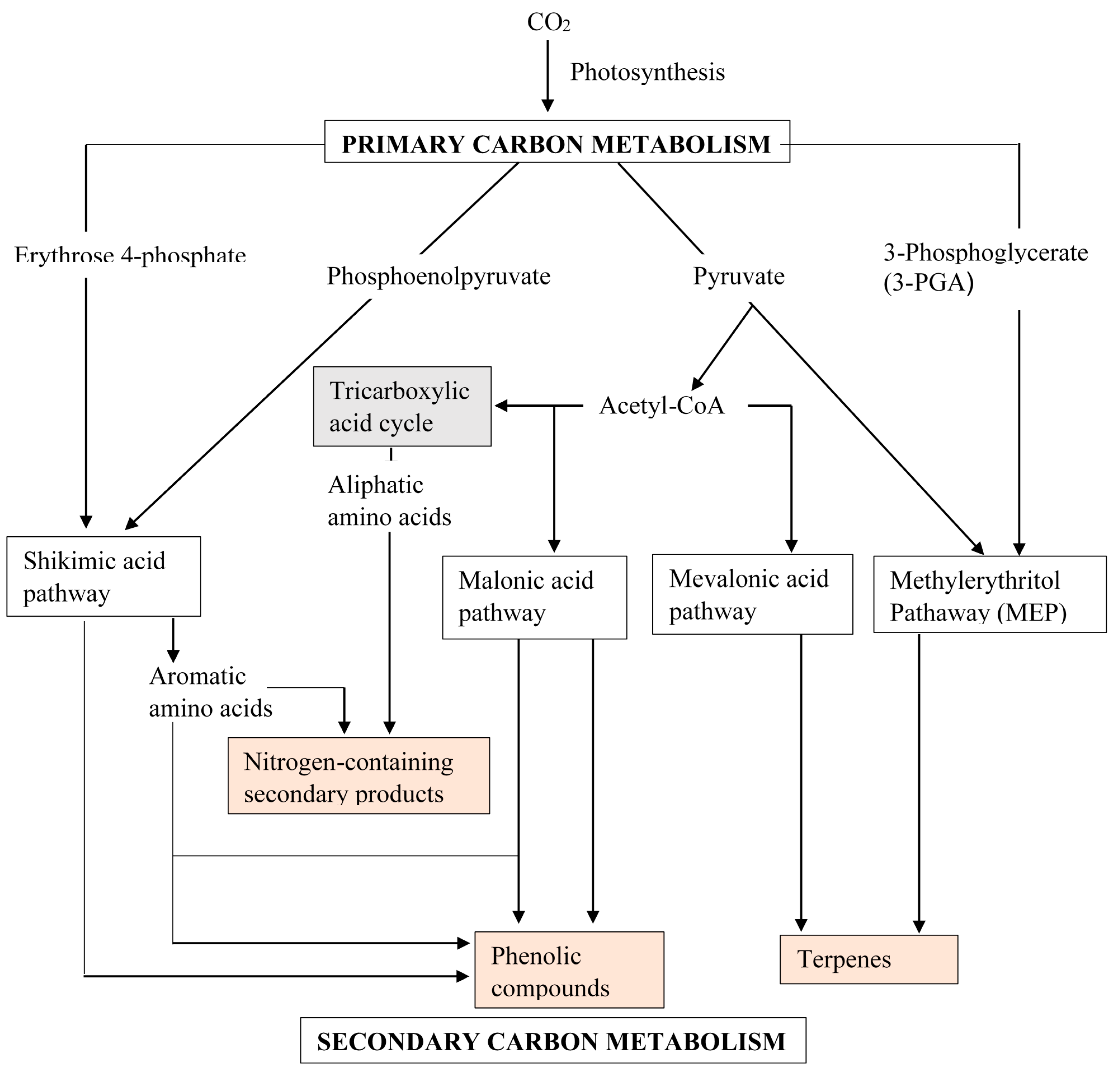
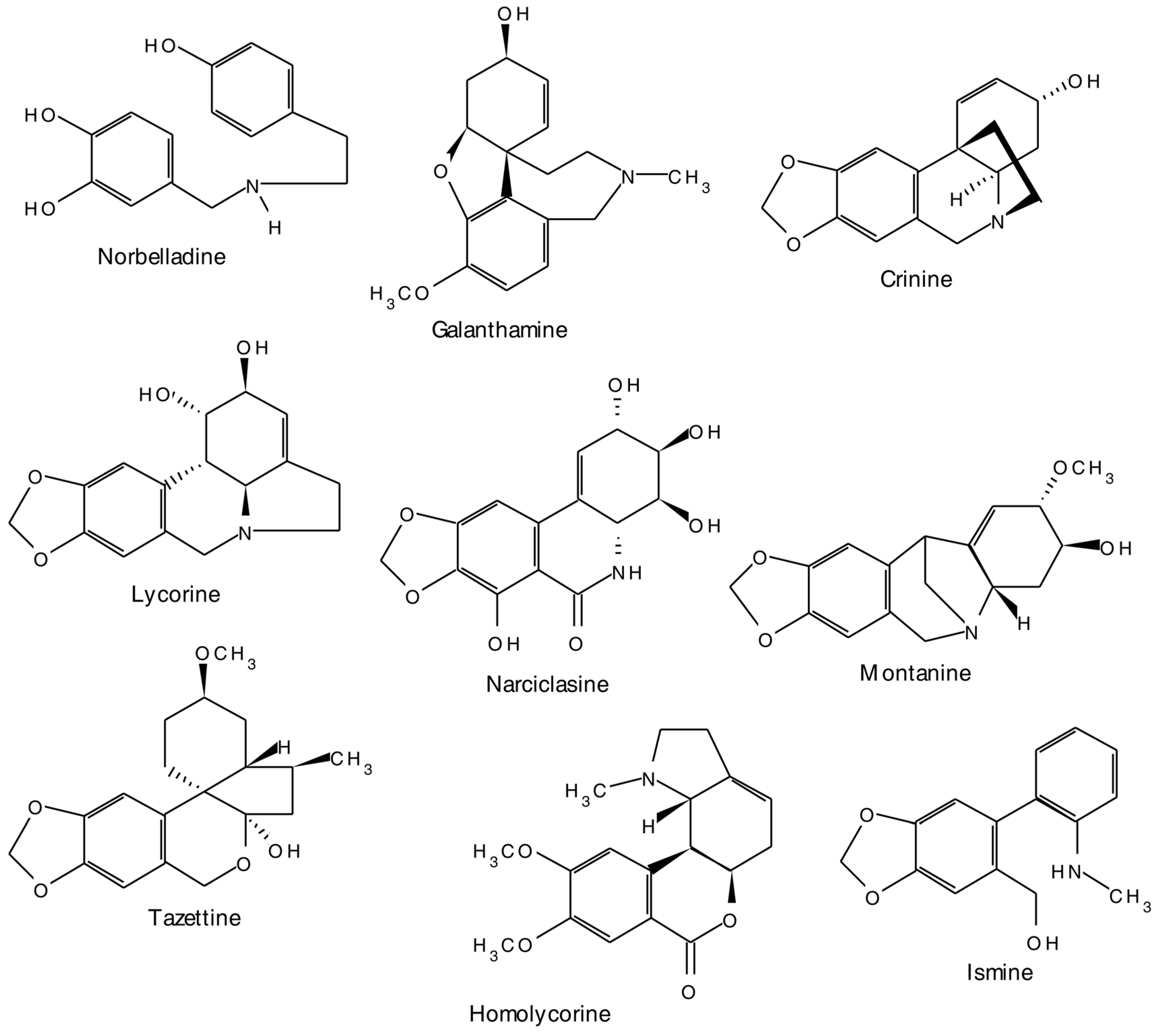
2.1. Alkaloids
| Alkaloid | Plant Source | Pharmaceutical Use |
|---|---|---|
| Tropane and Nicotine | ||
| Atropine | Hyoscyamus niger | Anticholinergic, antidote to nerve gas poisoning [36] |
| Cocaine | Erythroxylon coca | Topical anaesthetic, potent central nervous system stimulant, and adrenergic blocking agent; drug of abuse [30,37] |
| Codeine | Papaver somniferum | A nonaddictive analgesic and antitussive [30] |
| Morphine | Papaver somniferum | Powerful narcotic analgesic, addictive drug of abuse [30] |
| Nicotine | Nicotiana tabacum | Highly toxic, causes respiratory paralysis, horticultural insecticide; drug of abuse [37] |
| Scopolamine | Hyoscyamu. niger | Powerful narcotic, used as a sedative for motion sickness [36] |
| (+)-Tubocurarine | Chondrodendron tomentosm | Nondepolarising muscle relaxant producing paralysis, used as an adjuvant to anaesthesia [37] |
| Amarryllidaceae | ||
| Galanthamine | Galanthus woronowii | Used in the treatment of Alzheimer’s disease [37] |
| Piperidine | ||
| Coniine | Conium maculatum | An extremely toxic alkaloid, causes paralysis of motor nerve endings, used in homeopathy in minute doses [38] |
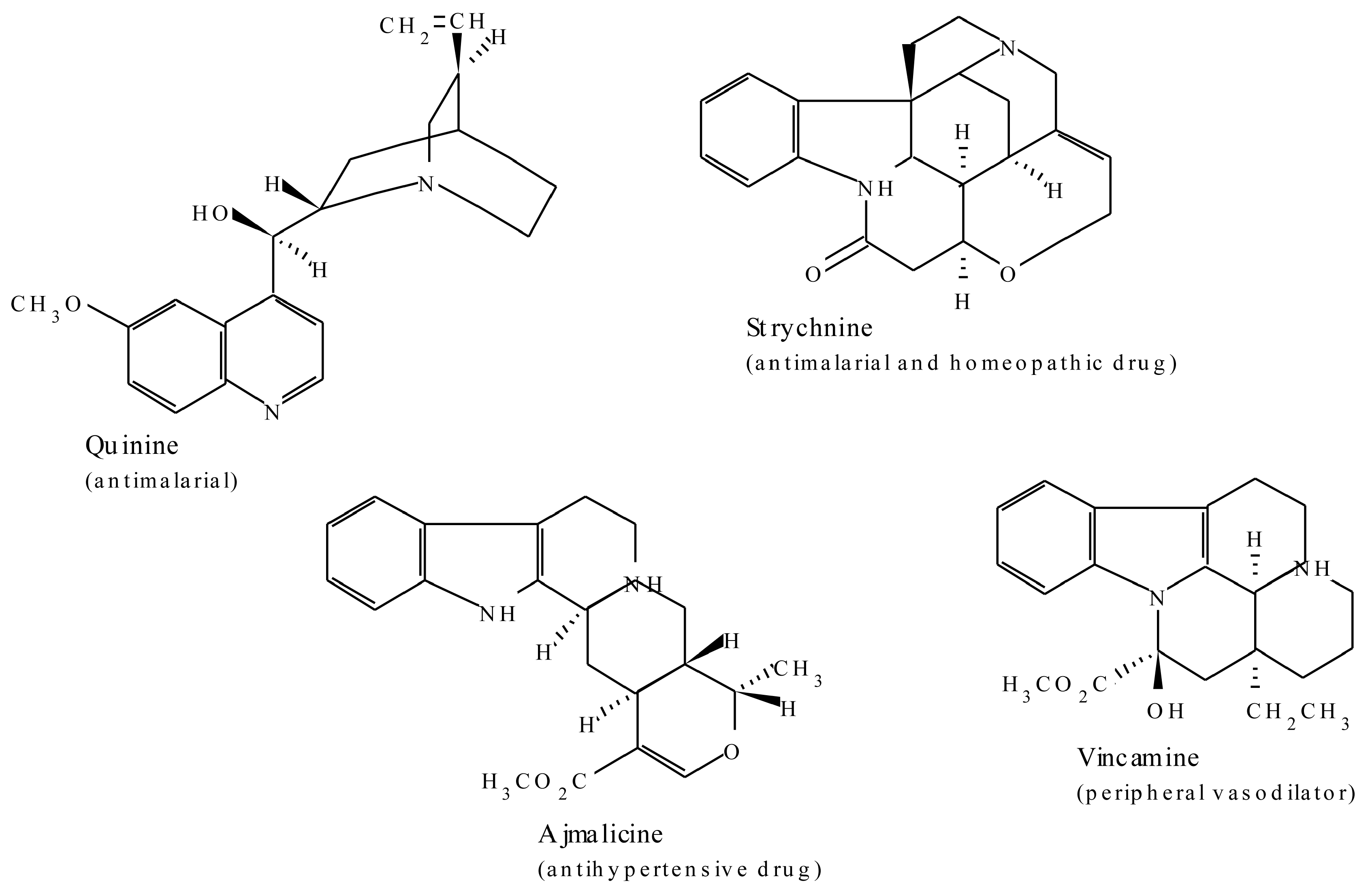
| Terpenoid Indole Alkaloids | ||
| Ajmaline | Rauwolfia serpentina | Antiarrythmic that functions by inhibiting glucose uptake by heart tissue mitochondria [37] |
| Camptothecin | Camptotheca acuminata | Potent anticancer agent [39,40] |
| Quinine | Cinchona officinalis | Traditional antimalarial, important in treating Plasmodium falciparum strains that are resistant to other antimalarials [41] |
| Strychnine | Strychnos nuxvomica | Tetanic poison, rat poison, used in homeopathy [37] |
| Vinblastine | Catharanthus roseus | Antineoplastic used to treat Hodgkin’s disease and other lymphomas |
| Vincristine | Catharanthus roseus | Am antitumor and chemotherapeutic agent [37] |
| Purine | ||
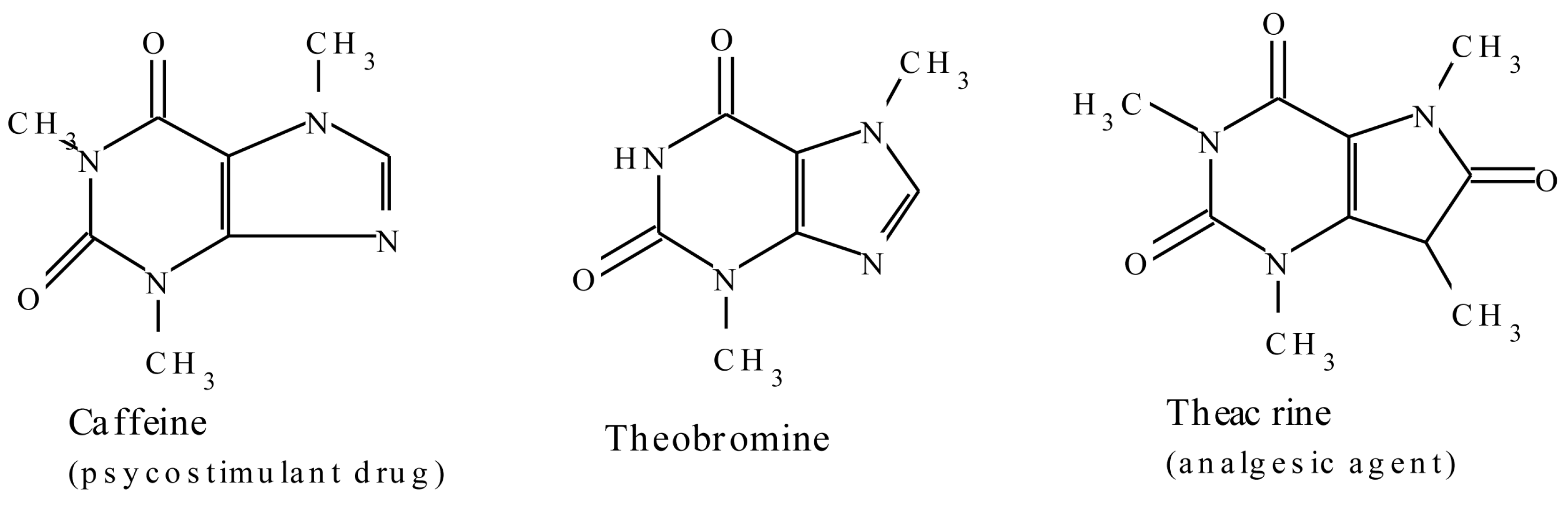
| Caffeine | Coffea arabica | Used as a central nervous system stimulant [42,43,44] |
| Pilocarpus | ||
| Pilocarpine | Pilocarpus jaborandi | Peripheral stimulant of the parasympathetic system, used to treat glaucoma [37] |
| Ipecac | ||
| Emetine | Uragoga ipecacuanha | Orally active emetic, amoebicide [37] |
| Benzophenanthridine | ||
| Sanguinarine | Eschscholzia californica | Antibacterial showing antiplaque activity, used in toothpastes and oral rinses [37] |
2.1.1. Tropane and Nicotine Alkaloids
2.1.2. Amaryllidaceae Alkaloids

2.1.3. Terpenoid Indole Alkaloids
2.1.4. Purine Alkaloids
2.2. Phenolic Compounds
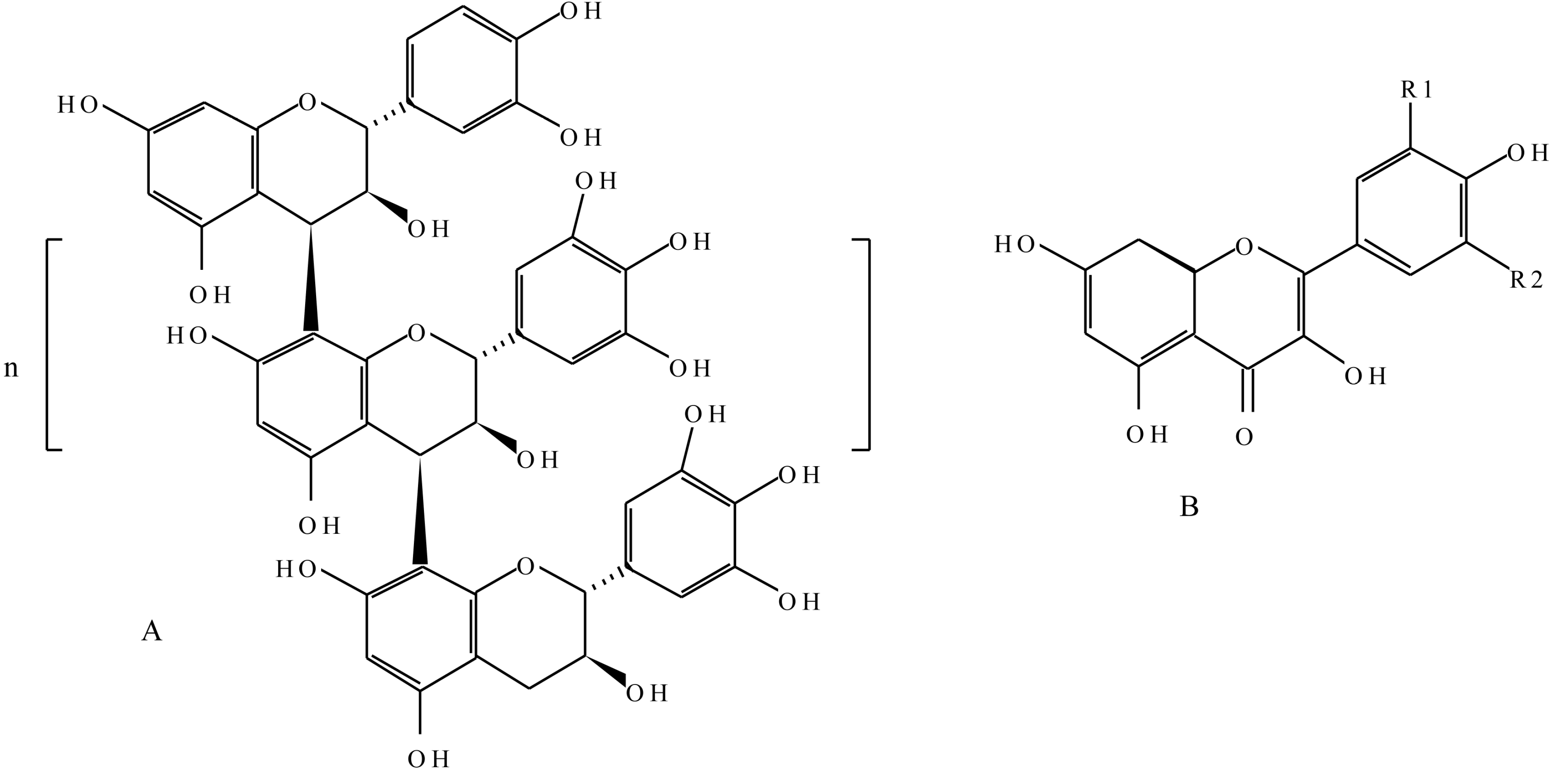
2.2.1. Flavonoids
2.2.2. Tannins
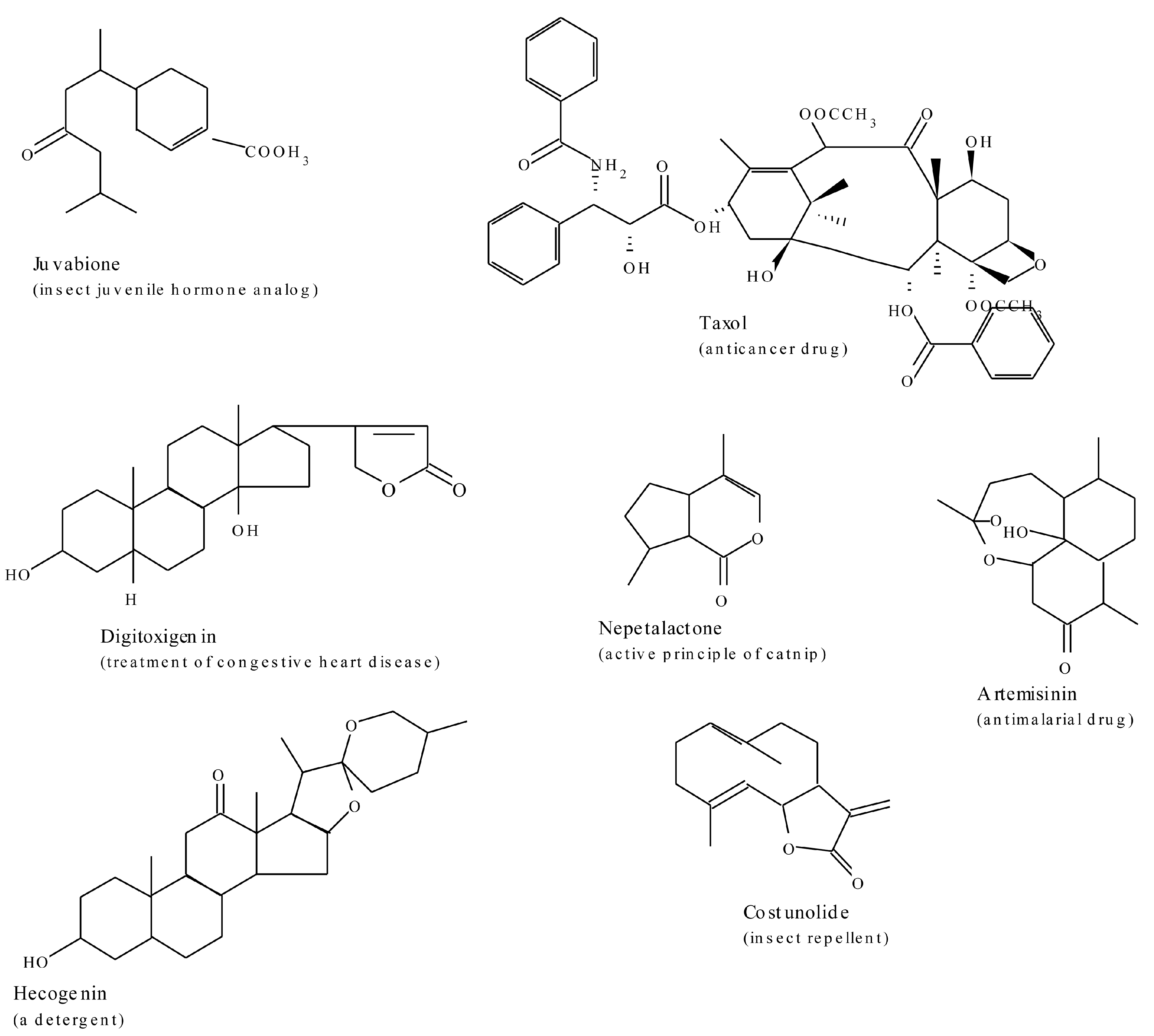
2.3. Terpenes
3. Metabolic Alterations
3.1. Metabolic Pathway Manipulations
3.2. Other Manipulated Factors
| Metabolite | Plant Species | Factor Manipulated | Ref. |
|---|---|---|---|
| Morphine, codeine | Papaver somniferum | Vertcillium dahliae | [136] |
| Indole alkaloids | Catharanthus roseus | Fungal elicitor | [133,137] |
| Indole alkaloids | Catharanthus roseus | Diethyl amino ethyl dichloro phenyl ether | [138] |
| N-acetyl-tryptamine | Catharanthus roseus | Pythium aphanidermatum | [139] |
| Ajmalicine | Catharanthus roseus | Trichoderma viride | [140] |
| Catharanthine | Catharanthus roseus | Vanadium sulphate | [141] |
| Camalexin, indole glucosinolates | Arabidopsis thaliana | Erwinia carotovora | [142] |
| Camalexin | Arabidopsis thaliana | Oxidative stress, amino acid starvation | [143] |
| Diterpenoid tanshinones | Salvia miltiorrhiza | Yeast elicitor | [144] |
| Rutacridone epoxide | Ruta graveolens | Chitosan | [145] |
| Silymarin | Silybum marianum | Yeast extract, Methyl jasmonate | [146] |
| Rosmarinic acid | Coleus blumei | [147] | |
| Saponins | Panax ginseng | Low-energy ultrasound | [148] |
| Diosgenin | Dioscorea deltoida | Rhizopus arrhizus | [149] |
| Hyoscyamine, scopolamine | Hyoscyamus niger | Fungal elicitor, | [150] |
| Hyoscyamine, scopolamine | Hyoscyamus muticus | Fungal elicitor, Methyl jasmonate | [150] |
| Salidroside | Rhodiola sachalinensis | Aspergillus niger, Coriolus versicolor, Ganoderma lucidum | [151] |
| Sanguinarine | Papaver bracteatum | Dendryphion | [152] |
| Taxol | Taxus chinensis | Fungal elicitor | [153] |
| Tropane alkaloids | Brugmansia suaveolens | Spodoptera frugiperda, Methyl jasmonate | [154] |
| Scopoletin | Ammi majus | Enterobacter sakazaki | [155] |
| Tanshinone | Salvia miltiorrhiza | Hyperosmotic stress, yeast elicitor | [156] |
| Acridone expoxide | Ruta gravelones | Fungal poly saccharide | [145] |
| Colchicine | Valeriana wallichii | Valepotriates | [157] |
| Sesquiterpenoids | Datura stramonium | Metal ions | [158] |
| Capsidiol, debneyol, scopoletin, nicotine | Nicotiana tabacum | Phytopthora cryptogea, Yeast extract, Cryptogein Cellulase, Methyl jasmonate | [159,160,161] |
| Raucaffrincine | Rauwolfia canescens | Yeast elicitor, Methyl jasmonate | [162,163] |
| Kinobeon A | Carthamus tinctorius | Blue green algae | [164] |
| Isoflavonoids | Lotus corniculatus | Glutathione | [165] |
| Digoxin, Purpureaglycoside A | Digitalis lanata | Temperature | [166] |
| Ubiquinone | Nicotiana tabacum | Temperature | [167] |
| Crude alkaloids | Catharanthus roseus | Temperature | [168] |
| Anthocyanin | Daucas carota | Light | [169] |
| Sesquiterpenes | Marticaria chamomilla | Light | [170] |
| Monoterpenes | Citrus limo | Light | [171] |
| Catechin, epicatechin, | Malus × domestica Borkh | Chilling | [172] |
| Flavonoids, tannins | Cyrtanthus contractus, C. guthrieae | Sodium chloride | [173] |
| Digitoxin | Digitalis purpurea | Phosphate | [174] |
| Betacyanin | Chenopodium rubrum | Phosphate | [174,175] |
| Betacyanin | Phytolacca americana | Phosphate | [38,176] |
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Noel, J.P.; Austin, M.B.; Bomati, E.K. Structure–function relationships in plant phenylpropanoid biosynthesis. Curr. Opin. Plant Biol. 2005, 8, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Ncube, B.; Finnie, J.; Van Staden, J. Quality from the field: The impact of environmental factors as quality determinants in medicinal plants. S. Afr. J. Bot. 2012, 82, 11–20. [Google Scholar] [CrossRef]
- Landry, L.G.; Chapple, C.C.; Last, R.L. Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol. 1995, 109, 1159–1166. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.E.; Rashotte, A.M.; Murphy, A.S.; Normanly, J.; Tague, B.W.; Peer, W.A.; Taiz, L.; Muday, G.K. Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol. 2001, 126, 524–535. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, T. Diversity and variability of plant secondary metabolism: A mechanistic view. Entomol. Exp. Appl. 1996, 80, 177–188. [Google Scholar] [CrossRef]
- Van der Fits, L.; Zhang, H.; Menke, F.L.; Deneka, M.; Memelink, J. A Catharanthus roseus BPF-1 homologue interacts with an elicitor-responsive region of the secondary metabolite biosynthetic gene Str and is induced by elicitor via a JA-independent signal transduction pathway. Plant Mol. Biol. 2000, 44, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Wink, M. Evolution of secondary metabolites from an ecological and molecular phylogenetic perspective. Phytochemistry 2003, 64, 3–19. [Google Scholar] [CrossRef]
- Benderoth, M.; Textor, S.; Windsor, A.J.; Mitchell-Olds, T.; Gershenzon, J.; Kroymann, J. Positive selection driving diversification in plant secondary metabolism. Proc. Natl. Acad. Sci. USA 2006, 103, 9118–9123. [Google Scholar] [CrossRef] [PubMed]
- Janzen, D.H. Host plants as islands in evolutionary and contemporary time. Am. Nat. 1968, 102, 592–595. [Google Scholar] [CrossRef]
- Pichersky, E.; Gang, D.R. Genetics and biochemistry of secondary metabolites in plants: An evolutionary perspective. Trends Plant Sci. 2000, 5, 439–445. [Google Scholar] [CrossRef]
- Hartmann, T.; Kutchan, T.M.; Strack, D. Evolution of metabolic diversity. Phytochemistry 2005, 66, 1198–1199. [Google Scholar] [CrossRef] [PubMed]
- Aharoni, A.; Galili, G. Metabolic engineering of the plant primary–secondary metabolism interface. Curr. Opin. Biotechnol. 2011, 22, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007, 50, 660–677. [Google Scholar] [CrossRef] [PubMed]
- Malitsky, S.; Blum, E.; Less, H.; Venger, I.; Elbaz, M.; Morin, S.; Eshed, Y.; Aharoni, A. The transcript and metabolite networks affected by the two clades of Arabidopsis glucosinolate biosynthesis regulators. Plant Physiol. 2008, 148, 2021–2049. [Google Scholar] [CrossRef] [PubMed]
- Frerigmann, H.; Berger, B.; Gigolashvili, T. bHLH05 is an interaction partner of MYB51 and a novel regulator of glucosinolate biosynthesis in Arabidopsis. Plant Physiol. 2014, 166, 349–369. [Google Scholar] [CrossRef] [PubMed]
- Maruyama-Nakashita, A.; Nakamura, Y.; Tohge, T.; Saito, K.; Takahashi, H. Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 2006, 18, 3235–3251. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Gaudinier, A.; Tang, M.; Taylor-Teeples, M.; Nham, N.T.; Ghaffari, C.; Benson, D.S.; Steinmann, M.; Gray, J.A.; Brady, S.M.; Kliebenstein, D.J. Promoter-based integration in plant defense regulation. Plant Physiol. 2014, 166, 1803–1820. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Grubb, C.D.; Abel, S. Glucosinolate metabolism and its control. Trends in Plant Sci. 2006, 11, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Henkes, S.; Sonnewald, U.; Badur, R.; Flachmann, R.; Stitt, M. A small decrease of plastid transketolase activity in antisense tobacco transformants has dramatic effects on photosynthesis and phenylpropanoid metabolism. Plant Cell 2001, 13, 535–551. [Google Scholar] [CrossRef] [PubMed]
- Tamagnone, L.; Merida, A.; Parr, A.; Mackay, S.; Culianez-Macia, F.A.; Roberts, K.; Martin, C. The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 1998, 10, 135–154. [Google Scholar] [CrossRef] [PubMed]
- Shufflebottom, D.; Edwards, K.; Schuch, W.; Bevan, M. Transcription of two members of a gene family encoding phenylalanine ammonia-lyase leads to remarkably different cell specificities and induction patterns. Plant J. 1993, 3, 835–845. [Google Scholar] [CrossRef] [PubMed]
- Weisshaar, B.; Jenkins, G.I. Phenylpropanoid biosynthesis and its regulation. Curr. Opin. Plant Biol. 1998, 1, 251–257. [Google Scholar] [CrossRef]
- Martin, C.; Paz-Ares, J. MYB transcription factors in plants. Trends Genet. 1997, 13, 67–73. [Google Scholar] [CrossRef]
- Van der Fits, L.; Memelink, J. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 2000, 289, 295–297. [Google Scholar] [CrossRef] [PubMed]
- Mol, J.; Grotewold, E.; Koes, R. How genes paint flowers and seeds. Trends Plant Sci. 1998, 3, 212–217. [Google Scholar] [CrossRef]
- Celenza, J.L.; Quiel, J.A.; Smolen, G.A.; Merrikh, H.; Silvestro, A.R.; Normanly, J.; Bender, J. The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol. 2005, 137, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, J.; Facchini, P.J. Alkaloid biosynthesis: Metabolism and trafficking. Annu. Rev. Plant Biol. 2008, 59, 735–769. [Google Scholar] [CrossRef] [PubMed]
- Fischbach, M.A.; Clardy, J. One pathway, many products. Nat. Chem. Biol. 2007, 3, 353–355. [Google Scholar] [CrossRef] [PubMed]
- Facchini, P.J. Plant secondary metabolism: Out of the evolutionary abyss. Trends Plant Sci. 1999, 4, 382–384. [Google Scholar] [CrossRef]
- De Luca, V.; St Pierre, B. The cell and developmental biology of alkaloid biosynthesis. Trends Plant Sci. 2000, 5, 168–173. [Google Scholar] [CrossRef]
- Facchini, P.J. Alkaloid biosynthesis in plants: Biochemistry, cell biology, molecular regulation, and metabolic engineering applications. Ann. Rev. Plant Biol. 2001, 52, 29–66. [Google Scholar] [CrossRef] [PubMed]
- Maplestone, R.A.; Stone, M.J.; Williams, D.H. The evolutionary role of secondary metabolites-a review. Gene 1992, 115, 151–157. [Google Scholar] [CrossRef]
- Kim, Y.S.; Sano, H. Pathogen resistance of transgenic tobacco plants producing caffeine. Phytochemistry 2008, 69, 882–888. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Uefuji, H.; Ogita, S.; Sano, H. Transgenic tobacco plants producing caffeine: A potential new strategy for insect pest control. Transgenic Res. 2006, 15, 667–672. [Google Scholar] [CrossRef] [PubMed]
- Kutchan, T.M. Alkaloid Biosynthesis [mdash] The basis for metabolic engineering of medicinal plants. Plant Cell 1995, 7, 1059. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.S. The Amaryllidaceae Alkaloids. In The Alkaloids: Chemistry and Phamarcology; Brossi, A., Ed.; Academic Press: London, UK, 1987; Volume 30. [Google Scholar]
- Vetter, J. Poison hemlock (Conium maculatum L.). Food Chem. Toxic. 2004, 42, 1373–1382. [Google Scholar] [CrossRef] [PubMed]
- Kjeldsen, E.; Svejstrup, J.Q.; Gromova, I.I.; Alsner, J.; Westergaard, O. Camptothecin inhibits both the cleavage and religation reactions of eukaryotic DNA topoisomerase I. J. Mol. Biol. 1992, 228, 1025–1030. [Google Scholar] [CrossRef]
- Liu, L.F.; Desai, S.D.; LI, T.K.; Mao, Y.; Sun, M.; SIM, S.P. Mechanism of action of camptothecin. Ann. N. Y. Acad. Sci. 2000, 922, 1–10. [Google Scholar] [CrossRef] [PubMed]
- López-Meyer, M.; Nessler, C.L. Tryptophan decarboxylase is encoded by two autonomously regulated genes in Camptotheca acuminata which are differentially expressed during development and stress. Plant J. 1997, 11, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Cauli, O.; Morelli, M. Caffeine and the dopaminergic system. Behav. Pharmacol. 2005, 16, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Fredholm, B.B.; Bättig, K.; Holmén, J.; Nehlig, A.; Zvartau, E.E. Actions of caffeine in the brain with special reference to factors that contribute to its widespread use. Pharmacol. Rev. 1999, 51, 83–133. [Google Scholar] [PubMed]
- Howell, L.; Coffin, V.L.; Spealman, R.D. Behavioral and physiological effects of xanthines in nonhuman primates. Psychopharmacology 1997, 129, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, T.; Yamada, Y. Alkaloid biogenesis: Molecular aspects. Ann. Rev. Plant Biol. 1994, 45, 257–285. [Google Scholar] [CrossRef]
- Nakajima, K.; Hashimoto, T.; Yamada, Y. Opposite stereospecificity of two tropinone reductases is conferred by the substrate-binding sites. J. Biol. Chem. 1994, 269, 11695–11698. [Google Scholar] [PubMed]
- Bentley, K.W. The Isoquinoline Alkaloids; CRC Press: London, UK, 1998. [Google Scholar]
- Unver, N. New skeletons and new concepts in Amaryllidaceae alkaloids. Phytochem. Rev. 2007, 6, 125–135. [Google Scholar] [CrossRef]
- Facchini, P.J.; de Luca, V. Phloem-specific expression of tyrosine/dopa decarboxylase genes and the biosynthesis of isoquinoline alkaloids in Opium poppy. Plant Cell 1995, 7, 1811–1821. [Google Scholar] [CrossRef] [PubMed]
- Chou, W.M.; Kutchan, T.M. Enzymatic oxidations in the biosynthesis of complex alkaloids. Plant J. 1998, 15, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Viladomat, F.; Bastida, J.; Codina, C.; Nair, J.J.; Campbell, W.E. Alkaloids of the South African Amaryllidaceae. In Recent Research Developments in Phytochemistry; Pandalai, S.G., Ed.; Research Signpost Publishers: Trivandrum, India, 1997; Volume 1. [Google Scholar]
- Houghton, P.J.; Ren, Y.; Howes, M.J. Acetylcholinesterase inhibitors from plants and fungi. Nat. Prod. Rep. 2006, 23, 181–199. [Google Scholar] [CrossRef] [PubMed]
- McNulty, J.; Thorat, A.; Vurgun, N.; Nair, J.J.; Makaji, E.; Crankshaw, D.J.; Holloway, A.C.; Pandey, S. Human cytochrome P450 liability studies of trans-dihydronarciclasine: A readily available, potent, and selective cancer cell growth inhibitor. J. Nat. Prod. 2010, 74, 106–108. [Google Scholar] [CrossRef] [PubMed]
- Nair, J.J.; van Staden, J. Pharmacological and toxicological insights to the South African Amaryllidaceae. Food Chem. Toxicol. 2013, 62, 262–275. [Google Scholar] [CrossRef] [PubMed]
- Pettit, G.R.; Pettit III, G.R.; Backhaus, R.A.; Boyd, M.R.; Meerow, A.W. Antineoplastic agents, 256. Cell growth inhibitory isocarbostyrils from Hymenocallis. J. Nat. Prod. 1993, 56, 1682–1687. [Google Scholar] [CrossRef] [PubMed]
- McLachlan, A.; Kekre, N.; McNulty, J.; Pandey, S. Pancratistatin: A natural anti-cancer compound that targets mitochondria specifically in cancer cells to induce apoptosis. Apoptosis 2005, 10, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.; Kekre, N.; Naderi, J.; McNulty, J. Induction of apoptotic cell death specifically in rat and human cancer cells by pancratistatin. Artif. Cells Nanomed. Biotechnol. 2005, 33, 279–295. [Google Scholar] [CrossRef]
- Lamoral-Theys, D.; Andolfi, A.; Van Goietsenoven, G.; Cimmino, A.; le Calvé, B.; Wauthoz, N.; Mégalizzi, V.; Gras, T.; Bruyère, C.; Dubois, J. Lycorine, the main phenanthridine Amaryllidaceae alkaloid, exhibits significant antitumor activity in cancer cells that display resistance to proapoptotic stimuli: An investigation of structure−activity relationship and mechanistic insight. J. Med. Chem. 2009, 52, 6244–6256. [Google Scholar] [CrossRef] [PubMed]
- Lefranc, F.; Sauvage, S.; Van Goietsenoven, G.; Mégalizzi, V.; Lamoral-Theys, D.; Debeir, O.; Spiegl-Kreinecker, S.; Berger, W.; Mathieu, V.; Decaestecker, C. Narciclasine, a plant growth modulator, activates Rho and stress fibers in glioblastoma cells. Mol. Cancer Ther. 2009, 8, 1739–1750. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Katano, N.; Ooi, A.; Inoue, K. Secologanin synthase which catalyzes the oxidative cleavage of loganin into secologanin is a cytochrome P450. Phytochemistry 2000, 53, 7–12. [Google Scholar] [CrossRef]
- Contin, A.; van der Heijden, R.; Lefeber, A.W.; Verpoorte, R. The iridoid glucoside secologanin is derived from the novel triose phosphate/pyruvate pathway in a Catharanthus roseus cell culture. FEBS Lett. 1998, 434, 413–416. [Google Scholar] [CrossRef] [PubMed]
- Priel, E.; Showalter, S.D.; Blair, D.G. Inhibition of human immunodeficiency virus (HTV-1) replication in vitro by noncytotoxic doses of camptothecin, a topoisomerase I inhibitor. AIDS Res. Hum. Retrovir. 1991, 7, 65–72. [Google Scholar] [PubMed]
- Bodley, A.L.; Shapiro, T.A. Molecular and cytotoxic effects of camptothecin, a topoisomerase I inhibitor, on trypanosomes and Leishmania. Proc. Natl. Acad. Sci. USA 1995, 92, 3726–3730. [Google Scholar] [CrossRef] [PubMed]
- Ashihara, H.; Monteiro, A.M.; Gillies, F.M.; Crozier, A. Biosynthesis of caffeine in leaves of coffee. Plant Physiology 1996, 111, 747–753. [Google Scholar] [PubMed]
- McCarthy, A.A.; McCarthy, J.G. The structure of two N-methyltransferases from the caffeine biosynthetic pathway. Plant Physiol. 2007, 144, 879–889. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Kanehara, T.; Shimizu, H.; Suzuki, T.; Gillies, F.M.; Crozier, A.; Ashihara, H. Caffeine biosynthesis in young leaves of Camellia sinensis: In vitro studies on N-methyltransferase activity involved in the conversion of xanthosine to caffeine. Physiol. Plant. 1996, 98, 629–636. [Google Scholar] [CrossRef]
- Nazario, G.M.; Lovatt, C.J. Separate de novo and salvage purine pools are involved in the biosynthesis of theobromine but not caffeine in leaves of Coffea arabica L. Plant Physiol. 1993, 103, 1203–1210. [Google Scholar] [PubMed]
- Porkka-Heiskanen, T.; Strecker, R.E.; Thakkar, M.; Bjørkum, A.A.; Greene, R.W.; McCarley, R.W. Adenosine: A mediator of the sleep-inducing effects of prolonged wakefulness. Science 1997, 276, 1265–1268. [Google Scholar] [CrossRef] [PubMed]
- Robertson, D.; Wade, D.; Workman, R.; Woosley, R.L.; Oates, J. Tolerance to the humoral and hemodynamic effects of caffeine in man. J. Clin. Investig. 1981, 67, 1111–1117. [Google Scholar] [CrossRef] [PubMed]
- Bravo, L. Polyphenols: Chemistry, dietary sources, metabolism, and nutritional significance. Nutr. Rev. 1998, 56, 317–333. [Google Scholar] [CrossRef] [PubMed]
- Dey, P.M.; Harborne, J.B. Methods in Plant Biochemistry: Plant Phenolics; Academic Press: New York, NY, USA, 1989; Volume 1. [Google Scholar]
- Ryan, D.; Robards, K.; Prenzler, P.; Antolovich, M. Applications of mass spectrometry to plant phenols. TrAC Trends Anal. Chem. 1999, 18, 362–372. [Google Scholar] [CrossRef]
- Dixon, R.A.; Paiva, N.L. Stress-induced phenylpropanoid metabolism. Plant Cell 1995, 7, 1085. [Google Scholar] [CrossRef] [PubMed]
- Crozier, A.; Jaganath, I.B.; Clifford, M.N. Phenols, polyphenols and tannins: An overview. In Plant Secondary Metabolites-Occurrence, Structure and Role in the Diet; Crozier, A., Clifford, M.N., Ashihara, H., Eds.; Blackwell Publishing: Oxford, UK, 2006. [Google Scholar]
- Rice-Evans, C.; Miller, N.; Paganga, G. Antioxidant properties of phenolic compounds. Trends Plant Sci. 1997, 2, 152–159. [Google Scholar] [CrossRef]
- Heim, K.E.; Tagliaferro, A.R.; Bobilya, D.J. Flavonoid antioxidants: chemistry, metabolism and structure-activity relationships. J. Nutr. Biochem. 2002, 13, 572–584. [Google Scholar] [CrossRef]
- Harborne, J.B.; Williams, C.A. Advances in flavonoid research since 1992. Phytochemistry 2000, 55, 481–504. [Google Scholar] [CrossRef]
- Waterman, P.G.; Mole, S. Analysis of Phenolic Plant Metabolites; Blackwell Scientific Publications: London, UK, 1994. [Google Scholar]
- Mori, A.; Nishino, C.; Enoki, N.; Tawata, S. Antibacterial activity and mode of action of plant flavonoids against Proteus vulgaris and Staphylococcus aureus. Phytochemistry 1987, 26, 2231–2234. [Google Scholar] [CrossRef]
- Kujumgiev, A.; Tsvetkova, I.; Serkedjieva, Y.; Bankova, V.; Christov, R.; Popov, S. Antibacterial, antifungal and antiviral activity of propolis of different geographic origin. J. Ethnopharmacol. 1999, 64, 235–240. [Google Scholar] [CrossRef]
- Burda, S.; Oleszek, W. Antioxidant and antiradical activities of flavonoids. J. Agric. Food Chem. 2001, 49, 2774–2779. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.A.; Harborne, J.B.; Geiger, H.; Hoult, J.R.S. The flavonoids of Tanacetum parthenium and T. vulgare and their anti-inflammatory properties. Phytochemistry 1999, 51, 417–423. [Google Scholar] [CrossRef]
- Middleton, E.; Kandaswami, C.; Theoharides, T.C. The effects of plant flavonoids on mammalian cells: Implications for inflammation, heart disease, and cancer. Pharmacol. Rev. 2000, 52, 673–751. [Google Scholar] [PubMed]
- Schofield, P.; Mbugua, D.; Pell, A. Analysis of condensed tannins: A review. Anim. Feed Sci. Technol. 2001, 91, 21–40. [Google Scholar] [CrossRef]
- Wallace, G.; Fry, S.C. Phenolic components of the plant cell wall. Int. Rev. Cytol. 1994, 151, 229–268. [Google Scholar] [PubMed]
- Haslam, E. Plant Polyphenols: Vegetable Tannins Revisited; Cambridge University Press: Melbourne, Australia, 1989. [Google Scholar]
- Mueller-Harvey, I. Analysis of hydrolysable tannins. Anim. Feed Sci. Technol. 2001, 91, 3–20. [Google Scholar] [CrossRef]
- Hagerman, A.E. Tannin Chemistry; Miami University: Washington, DC, USA, 2002. [Google Scholar]
- Xie, D.Y.; Dixon, R.A. Proanthocyanidin biosynthesis–still more questions than answers? Phytochemistry 2005, 66, 2127–2144. [Google Scholar] [CrossRef] [PubMed]
- Feeny, P. Seasonal changes in oak leaf tannins and nutrients as a cause of spring feeding by winter moth caterpillars. Ecology 1970, 51, 565–581. [Google Scholar] [CrossRef]
- Haslam, E. Natural polyphenols (vegetable tannins) as drugs: Possible modes of action. J. Nat. Prod. 1996, 59, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Gurib-Fakim, A. Medicinal plants: Traditions of yesterday and drugs of tomorrow. Mol. Asp. Med. 2006, 27, 1–93. [Google Scholar] [CrossRef] [PubMed]
- Okuda, T.; Yoshida, T.; Hatano, T. Pharmacologically Active Tannins Isolated From Medicinal Plants. In Plant Polyphenols: Synthesis, Properties, Significance; Hemingway, R.W., Laks, P.E., Eds.; Plenum Press: New York, NY, USA, 1992. [Google Scholar]
- Schultz, J.C.; Hunter, M.D.; Appel, H.M. Antimicrobial Activity of Polyphenols Mediates Plant-Herbivore Interactions. In Plant Polyphenols: Synthesis, Properties, Significance; Hemingway, R.W., Laks, P.E., Eds.; Plenum Press: New York, NY, USA, 1992. [Google Scholar]
- Vanden Berghe, D.A.; Vlietinck, A.J.; Van Hoof, L. Present Status and Prospects of Plant Products as Antiviral Agents. In Advances in Medicinal Plant Research; Vlietinck, A.J., Dommisse, R.A., Eds.; Wissenchaftliche Verlagsgesellschafte: Stuttgart, Germany, 1985. [Google Scholar]
- Loomis, W. [54] Overcoming problems of phenolics and quinones in the isolation of plant enzymes and organelles. Methods Enzymol. 1974, 31, 528–544. [Google Scholar] [PubMed]
- Bartram, S.; Jux, A.; Gleixner, G.; Boland, W. Dynamic pathway allocation in early terpenoid biosynthesis of stress-induced lima bean leaves. Phytochemistry 2006, 67, 1661–1672. [Google Scholar] [CrossRef] [PubMed]
- McGarvey, D.J.; Croteau, R. Terpenoid metabolism. Plant Cell 1995, 7, 1015. [Google Scholar] [CrossRef] [PubMed]
- Bick, J.A.; Lange, B.M. Metabolic cross talk between cytosolic and plastidial pathways of isoprenoid biosynthesis: unidirectional transport of intermediates across the chloroplast envelope membrane. Arch. Biochem. Biophys. 2003, 415, 146–154. [Google Scholar] [CrossRef]
- Laule, O.; Fürholz, A.; Chang, H.S.; Zhu, T.; Wang, X.; Heifetz, P.B.; Gruissem, W.; Lange, M. Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2003, 100, 6866–6871. [Google Scholar] [CrossRef] [PubMed]
- Zulak, K.G.; Bohlmann, J. Terpenoid biosynthesis and specialized vascular cells of conifer defense. J. Integr. Plant Biol. 2010, 52, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Bohlmann, J.; Keeling, C.I. Terpenoid biomaterials. Plant J. 2008, 54, 656–669. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.E. Toward a science of metabolic engineering. Science 1991, 252, 1668–1675. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.E.; Sburlati, A.; Hatzimanikatis, V.; Lee, K.; Renner, W.A.; Tsai, P.S. Inverse metabolic engineering: A strategy for directed genetic engineering of useful phenotypes. Biotechnol. Bioeng. 1996, 52, 109–121. [Google Scholar] [CrossRef]
- Sauer, U. Metabolic networks in motion: 13C-based flux analysis. Mol. Syst. Biol. 2006, 2. [Google Scholar] [CrossRef] [PubMed]
- Çakar, Z.P.; Turanlı-Yıldız, B.; Alkım, C.; Yılmaz, Ü. Evolutionary engineering of Saccharomyces cerevisiae for improved industrially important properties. FEMS Yeast Res. 2012, 12, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Aderem, A. Systems biology: its practice and challenges. Cell 2005, 121, 511–513. [Google Scholar] [CrossRef] [PubMed]
- Workman, C.T.; Mak, H.C.; McCuine, S.; Tagne, J.B.; Agarwal, M.; Ozier, O.; Begley, T.J.; Samson, L.D.; Ideker, T. A systems approach to mapping DNA damage response pathways. Science 2006, 312, 1054–1059. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.A. Engineering of plant natural product pathways. Curr. Opin. Plant Biol. 2005, 8, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Grotewold, E. Transcription factors for predictive plant metabolic engineering: Are we there yet? Curr. Opin. Biotechnol. 2008, 19, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Gerstein, M. Genomic analysis of the hierarchical structure of regulatory networks. Proc. Natl. Acad. Sci. USA 2006, 103, 14724–14731. [Google Scholar] [CrossRef] [PubMed]
- Hwang, E.I.; Kaneko, M.; Ohnishi, Y.; Horinouchi, S. Production of plant-specific flavanones by Escherichia coli containing an artificial gene cluster. Appl. Environ. Microbiol. 2003, 69, 2699–2706. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.V.; Armstrong, G.O.; Merwe, M.J.; Lambrechts, M.G.; Vivier, M.A.; Pretorius, I.S. Metabolic engineering of Saccharomyces cerevisiae for the synthesis of the wine-related antioxidant resveratrol. FEMS Yeast Res. 2003, 4, 79–85. [Google Scholar] [CrossRef]
- Martin, V.J.; Pitera, D.J.; Withers, S.T.; Newman, J.D.; Keasling, J.D. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 2003, 21, 796–802. [Google Scholar] [CrossRef] [PubMed]
- Steppuhn, A.; Gase, K.; Krock, B.; Halitschke, R.; Baldwin, I.T. Nicotine’s defensive function in nature. PLoS Biol. 2004, 2, e217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chintapakorn, Y.; Hamill, J.D. Antisense-mediated down-regulation of putrescine N-methyltransferase activity in transgenic Nicotiana tabacum L. can lead to elevated levels of anatabine at the expense of nicotine. Plant Mol. Biol. 2003, 53, 87–105. [Google Scholar] [CrossRef] [PubMed]
- Allen, R.S.; Millgate, A.G.; Chitty, J.A.; Thisleton, J.; Miller, J.A.; Fist, A.J.; Gerlach, W.L.; Larkin, P.J. RNAi-mediated replacement of morphine with the nonnarcotic alkaloid reticuline in opium poppy. Nat. Biotechnol. 2004, 22, 1559–1566. [Google Scholar] [CrossRef] [PubMed]
- Yun, D.J.; Hashimoto, T.; Yamada, Y. Metabolic engineering of medicinal plants: transgenic Atropa belladonna with an improved alkaloid composition. Proc. Natl. Acad. Sci. USA 1992, 89, 11799–11803. [Google Scholar] [CrossRef] [PubMed]
- Croteau, R.; Kutchan, T.M.; Lewis, N.G. Natural Products (Secondary Metabolites). In Biochemistry and Molecular Biology of Plants; Buchanan, B.B., Gruissem, W., Jones, R.L., Eds.; American Society of Plant Physiologists: Rockville, MD, USA, 2000; Volume 40. [Google Scholar]
- Yu, O.; Jung, W.; Shi, J.; Croes, R.A.; Fader, G.M.; McGonigle, B.; Odell, J.T. Production of the isoflavones genistein and daidzein in non-legume dicot and monocot tissues. Plant Physiol. 2000, 124, 781–794. [Google Scholar] [CrossRef] [PubMed]
- Bruce, W.; Folkerts, O.; Garnaat, C.; Crasta, O.; Roth, B.; Bowen, B. Expression profiling of the maize flavonoid pathway genes controlled by estradiol-inducible transcription factors CRC and P. Plant Cell 2000, 12, 65–79. [Google Scholar] [CrossRef] [PubMed]
- Yu, O.; Shi, J.; Hession, A.O.; Maxwell, C.A.; McGonigle, B.; Odell, J.T. Metabolic engineering to increase isoflavone biosynthesis in soybean seed. Phytochemistry 2003, 63, 753–763. [Google Scholar] [CrossRef]
- Muir, S.R.; Collins, G.J.; Robinson, S.; Hughes, S.; Bovy, A.; de Vos, C.R.; van Tunen, A.J.; Verhoeyen, M.E. Overexpression of petunia chalcone isomerase in tomato results in fruit containing increased levels of flavonols. Nat. Biotechnol. 2001, 19, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Bovy, A.; de Vos, R.; Kemper, M.; Schijlen, E.; Pertejo, M.A.; Muir, S.; Collins, G.; Robinson, S.; Verhoeyen, M.; Hughes, S. High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes LC and C1. Plant Cell 2002, 14, 2509–2526. [Google Scholar] [CrossRef] [PubMed]
- Sommer, S.; Köhle, A.; Yazaki, K.; Shimomura, K.; Bechthold, A.; Heide, L. Genetic engineering of shikonin biosynthesis hairy root cultures of Lithospermum erythrorhizon transformed with the bacterial ubiC gene. Plant Mol. Biol. 1999, 39, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Jouhikainen, K.; Lindgren, L.; Jokelainen, T.; Hiltunen, R.; Teeri, T.H.; Oksman-Caldentey, K.M. Enhancement of scopolamine production in Hyoscyamus muticus L. hairy root cultures by genetic engineering. Planta 1999, 208, 545–551. [Google Scholar] [CrossRef]
- Sato, F.; Hashimoto, T.; Hachiya, A.; Tamura, K.I.; Choi, K.B.; Morishige, T.; Fujimoto, H.; Yamada, Y. Metabolic engineering of plant alkaloid biosynthesis. Proc. Natl. Acad. Sci. USA 2001, 98, 367–372. [Google Scholar] [CrossRef] [PubMed]
- Verpoorte, R.; Memelink, J. Engineering secondary metabolite production in plants. Curr. Opin. Biotechnol. 2002, 13, 181–187. [Google Scholar] [CrossRef]
- Ncube, B.; Finnie, J.F.; van Staden, J. Carbon–nitrogen ratio and in vitro assimilate partitioning patterns in Cyrtanthus guthrieae L. Plant Physiol. Biochem. 2014, 74, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.X.; Li, C.J.; Nan, Z.B. Effects of salt and drought stress on alkaloid production in endophyte-infected drunken horse grass (Achnatherum inebrians). Biochem. Syst. Ecol. 2011, 39, 471–476. [Google Scholar] [CrossRef]
- Glynn, C.; Rönnberg-Wästljung, A.C.; Julkunen-Tiitto, R.; Weih, M. Willow genotype, but not drought treatment, affects foliar phenolic concentrations and leaf-beetle resistance. Entomol. Exp. Appl. 2004, 113, 1–14. [Google Scholar] [CrossRef]
- Gouinguené, S.P.; Turlings, T.C. The effects of abiotic factors on induced volatile emissions in corn plants. Plant Physiol. 2002, 129, 1296–1307. [Google Scholar] [CrossRef] [PubMed]
- Eilert, U.; de Luca, V.; Constabel, F.; Kurz, W. Elicitor-mediated induction of tryptophan decarboxylase and strictosidine synthase activities in cell suspension cultures of Catharanthus roseus. Arch. Biochem. Biophys. 1987, 254, 491–497. [Google Scholar] [CrossRef]
- Pasquali, G.; Goddijn, O.J.; de Waal, A.; Verpoorte, R.; Schilperoort, R.A.; Hoge, J.H.C.; Memelink, J. Coordinated regulation of two indole alkaloid biosynthetic genes from Catharanthus roseus by auxin and elicitors. Plant Mol. Biol. 1992, 18, 1121–1131. [Google Scholar] [CrossRef] [PubMed]
- Menke, F.L.; Parchmann, S.; Mueller, M.J.; Kijne, J.W.; Memelink, J. Involvement of the octadecanoid pathway and protein phosphorylation in fungal elicitor-induced expression of terpenoid indole alkaloid biosynthetic genes in Catharanthus roseus. Plant Physiol. 1999, 119, 1289–1296. [Google Scholar] [CrossRef] [PubMed]
- Heinstein, P.F. Future approaches to the formation of secondary natural products in plant cell suspension cultures. J. Nat. Prod. 1985, 48, 1–9. [Google Scholar] [CrossRef]
- Rijhwani, S.K.; Shanks, J.V. Effect of elicitor dosage and exposure time on biosynthesis of indole alkaloids by Catharanthus roseus hairy root cultures. Biotechnol. Prog. 1998, 14, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.T.; Yamakawa, T.; Kodama, T.; Shimomura, K. Effects of chemicals on alkaloid production by transformed roots of belladonna. Phytochemistry 1998, 49, 2343–2347. [Google Scholar] [CrossRef]
- Eilert, U.; Constabel, F.; Kurz, W. Elicitor-stimulation of monoterpene indole alkaloid formation in suspension cultures of Catharanthus roseus. J. Plant Physiol. 1986, 126, 11–22. [Google Scholar] [CrossRef]
- Namdeo, A.; Patil, S.; Fulzele, D.P. Influence of fungal elicitors on production of ajmalicine by cell cultures of Catharanthus roseus. Biotechnol. Prog. 2002, 18, 159–162. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.; Smart, N.; Misawa, M.; Kurz, W.; Tallevi, S.; DiCosmo, F. Increased accumulation of indole alkaloids by some cell lines of Catharanthus roseus in response to addition of vanadyl sulphate. Plant Cell Rep. 1987, 6, 142–145. [Google Scholar] [PubMed]
- Brader, G.; Tas, É.; Palva, E.T. Jasmonate-dependent induction of indole glucosinolates in arabidopsis by culture filtrates of the nonspecific pathogenerwinia carotovora. Plant Physiol. 2001, 126, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Williams, C.C.; Last, R.L. Induction of Arabidopsis tryptophan pathway enzymes and camalexin by amino acid starvation, oxidative stress, and an abiotic elicitor. Plant Cell 1998, 10, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Hu, Z.; Tan, R.X.; Wu, J. Efficient production and recovery of diterpenoid tanshinones in Salvia miltiorrhiza hairy root cultures with in situ adsorption, elicitation and semi-continuous operation. J. Biotechnol. 2005, 119, 416–424. [Google Scholar] [CrossRef] [PubMed]
- Eilert, U.; Ehmke, A.; Wolters, B. Elicitor-Induced Accumulation of Acridone Alkaloid Epoxides in Ruta graveolens Suspension Cultures. Planta Med. 1984, 50, 508–512. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Sampedro, M.A.; Fernández-Tárrago, J.; Corchete, P. Yeast extract and methyl jasmonate-induced silymarin production in cell cultures of Silybum marianum (L.) Gaertn. J. Biotechnol. 2005, 119, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Szabo, E.; Thelen, A.; Petersen, M. Fungal elicitor preparations and methyl jasmonate enhance rosmarinic acid accumulation in suspension cultures of Coleus blumei. Plant Cell Rep. 1999, 18, 485–489. [Google Scholar] [CrossRef]
- Wu, J.; Lin, L. Elicitor-like effects of low-energy ultrasound on plant (Panax ginseng) cells: Induction of plant defense responses and secondary metabolite production. Appl. Microbiol. Biotechnol. 2002, 59, 51–57. [Google Scholar] [PubMed]
- Rokem, J.; Schwarzberg, J.; Goldberg, I. Autoclaved fungal mycelia increase diosgenin production in cell suspension cultures of Dioscorea deltoidea. Plant Cell Rep. 1984, 3, 159–160. [Google Scholar] [CrossRef] [PubMed]
- Singh, G. Fungal Elicitation of Plant Root Cultures-Application to Bioreactor Dosage. Ph.D. Thesis, Pennsylvania State University, USA, 1995. [Google Scholar]
- Zhou, X.; Wu, Y.; Wang, X.; Liu, B.; Xu, H. Salidroside production by hairy roots of Rhodiola sachalinensis obtained after transformation with Agrobacterium rhizogenes. Biol. Pharm. Bull. 2007, 30, 439–442. [Google Scholar] [CrossRef] [PubMed]
- Cline, S.D.; Coscia, C.J. Stimulation of sanguinarine production by combined fungal elicitation and hormonal deprivation in cell suspension cultures of Papaver bracteatum. Plant Physiol. 1988, 86, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wu, J.; Mei, X. Enhancement of taxol production and excretion in Taxus chinensis cell culture by fungal elicitation and medium renewal. App. Microbiol. Biotechnol. 2001, 55, 404–410. [Google Scholar] [CrossRef]
- Alves, M.N.; Sartoratto, A.; Trigo, J.R. Scopolamine in Brugmansia suaveolens (Solanaceae): Defense, allocation, costs, and induced response. J. Chem. Ecol. 2007, 33, 297–309. [Google Scholar] [CrossRef] [PubMed]
- Staniszewska, I.; Królicka, A.; Maliński, E.; Łojkowska, E.; Szafranek, J. Elicitation of secondary metabolites in in vitro cultures of Ammi majus L. Enzyme Microb. Technol. 2003, 33, 565–568. [Google Scholar] [CrossRef]
- Shi, M.; Kwok, K.; Wu, J.Y. Enhancement of tanshinone production in Salvia miltiorrhiza Bunge (red or Chinese sage) hairy-root culture by hyperosmotic stress and yeast elicitor. Biotechnol. Appl. Biochem. 2007, 46, 191–196. [Google Scholar] [PubMed]
- Becker, H.; Chavadej, S. Valepotriate production of normal and colchicine-treated cell suspension cultures of Valeriana wallichii. J. Nat. Prod. 1985, 48, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Threlfall, D.; Whitehead, I. The use of biotic and abiotic elicitors to induce the formation of secondary plant products in cell suspension cultures of solanaceous plants. Trans. Biochem. Soc. (UK) 1988, 16, 71–75. [Google Scholar]
- Lecourieux, D.; Mazars, C.; Pauly, N.; Ranjeva, R.; Pugin, A. Analysis and effects of cytosolic free calcium increases in response to elicitors in Nicotiana plumbaginifolia cells. Plant Cell 2002, 14, 2627–2641. [Google Scholar] [CrossRef] [PubMed]
- Milat, M.L.; Ricci, P.; Bonnet, P.; Blein, J.P. Capsidiol and ethylene production by tobacco cells in response to cryptogein, an elicitor from Phytophthora cryptogea. Phytochemistry 1991, 30, 2171–2173. [Google Scholar] [CrossRef]
- Wibberley, M.S.; Lenton, J.R.; Neill, S.J. Sesquiterpenoid phytoalexins produced by hairy roots of Nicotiana tabacum. Phytochemistry 1994, 37, 349–351. [Google Scholar] [CrossRef]
- Parchmann, S.; Gundlach, H.; Mueller, M.J. Induction of 12-oxo-phytodienoic acid in wounded plants and elicited plant cell cultures. Plant Physiol. 1997, 115, 1057–1064. [Google Scholar] [CrossRef] [PubMed]
- Gundlach, H.; Müller, M.J.; Kutchan, T.M.; Zenk, M.H. Jasmonic acid is a signal transducer in elicitor-induced plant cell cultures. Proc. Natl. Acad. Sci. USA 1992, 89, 2389–2393. [Google Scholar] [CrossRef] [PubMed]
- Hanagata, N.; Uehara, H.; Ito, A.; Takeuchi, T.; Karube, I. Elicitor for red pigment formation in Carthamus tinctorius cultured cells. J. Biotechnol. 1994, 34, 71–77. [Google Scholar] [CrossRef]
- Robbins, M.; Thomas, B.; Morris, P. Phenylpropanoid defence responses in transgenic Lotus corniculatus II. Modelling plant defence responses in transgenic root cultures using thiol and carbohydrate elicitors. J. Exp. Bot. 1995, 46, 513–524. [Google Scholar] [CrossRef]
- Kreis, W.; Reinhard, E. 12β-Hydroxylation of digitoxin by suspension-cultured Digitalis lanata cells: Production of digoxin in 20-litre and 300-litre air-lift bioreactors. J. Biotechnol. 1992, 26, 257–273. [Google Scholar] [CrossRef]
- Ikeda, T.; Matsumoto, T.; Noguchi, M. Effects of inorganic nitrogen sources and physical factors on the formation of ubiquinone by tobacco plant cells in suspension culture. Agric. Biol. Chem. 1977, 41, 1197–1201. [Google Scholar] [CrossRef]
- Courtois, D.; Guern, J. Temperature response of Catharanthus roseus cells cultivated in liquid medium. Plant Sci. Lett. 1980, 17, 473–482. [Google Scholar] [CrossRef]
- Gläßgen, W.E.; Wray, V.; Strack, D.; Metzger, J.W.; Seitz, H.U. Anthocyanins from cell suspension cultures of Daucus carota. Phytochemistry 1992, 31, 1593–1601. [Google Scholar] [CrossRef] [PubMed]
- Mulder-Krieger, T.; Verpoorte, R.; Svendsen, A.B.; Scheffer, J. Production of essential oils and flavours in plant cell and tissue cultures. A review. Plant Cell Tissue Org. Cult. 1988, 13, 85–154. [Google Scholar] [CrossRef]
- Leisso, R.S.; Buchanan, D.A.; Lee, J.; Mattheis, J.P.; Sater, C.; Hanrahan, I.; Watkins, C.B.; Gapper, N.; Johnston, J.W.; Schaffer, R.J. Chilling-related cell damage of apple (Malus× domestica Borkh.) fruit cortical tissue impacts antioxidant, lipid and phenolic metabolism. Physiol. Plant. 2015, 153, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Ncube, B.; Finnie, J.F.; Van Staden, J. Dissecting the stress metabolic alterations in in vitro Cyrtanthus regenerants. Plant Physiol. Biochem. 2013, 65, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Bohm, H.; Rink, E. Betalaines. Cell Cult. Somat. Cell Genet. Plant. 1988, 5, 449–463. [Google Scholar]
- Hagimori, M.; Matsumoto, T.; Obi, Y. Studies on the production of Digitalis cardenolides by plant tissue culture III. Effects of nutrients on digitoxin formation by shoot-forming cultures of Digitalis purpurea L. grown in liquid media. Plant Cell Physiol. 1982, 23, 1205–1211. [Google Scholar]
- Hagimori, M.; Matsumoto, T.; Obi, Y. Studies on the production of Digitalis cardenolides by plant tissue culture II. Effect of light and plant growth substances on digitoxin formation by Undifferentiated cells and shoot-forming cultures of Digitalis purpurea L. grown in liquid media. Plant Physiol. 1982, 69, 653–656. [Google Scholar] [CrossRef] [PubMed]
- Schliemann, W.; Joy, R.W.; Komamine, A.; Metzger, J.W.; Nimtz, M.; Wray, V.; Strack, D. Betacyanins from plants and cell cultures of Phytolacca americana. Phytochemistry 1996, 42, 1039–1046. [Google Scholar] [CrossRef]
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ncube, B.; Van Staden, J. Tilting Plant Metabolism for Improved Metabolite Biosynthesis and Enhanced Human Benefit. Molecules 2015, 20, 12698-12731. https://doi.org/10.3390/molecules200712698
Ncube B, Van Staden J. Tilting Plant Metabolism for Improved Metabolite Biosynthesis and Enhanced Human Benefit. Molecules. 2015; 20(7):12698-12731. https://doi.org/10.3390/molecules200712698
Chicago/Turabian StyleNcube, Bhekumthetho, and Johannes Van Staden. 2015. "Tilting Plant Metabolism for Improved Metabolite Biosynthesis and Enhanced Human Benefit" Molecules 20, no. 7: 12698-12731. https://doi.org/10.3390/molecules200712698