3.1. General Information
The reagents were purchased from Adamas-beta (Shanghai, China), J & K (Beijing, China), Energy Chemical (Shanghai, China) and TCI (Shanghai, China) and used without further purification. Analytical thin-layer chromatography (TLC) was performed on HSGF 254 plates (150–200 µm thickness, Yantai Huiyou Company, Yantai, China). Yields were not optimized. Melting points were measured in capillary tubes on a SGW X-4 melting point apparatus (Shanghai Jingmiyiqi Company, Shanghai, China) without correction. Nuclear magnetic resonance (NMR) spectra were recorded on a AVANCE 400 NMR (Bruker, Karlsruhe, Germany). Chemical shifts are reported in parts per million (ppm, δ) downfield from tetramethylsilane (TMS) used as the internal standard. Proton coupling patterns are described as singlet (s), doublet (d), triplet (t), quartet (q), multiplet (m), and broad (br). Low-and high-resolution mass spectra (LRMS and HRMS) were obtained with electric ionization (EI) and electrospray ionization (ESI) on Finnigan MAT-95 (Bremen, Germany) and LCQ-DECA (Bremen, Germany) spectrometers.
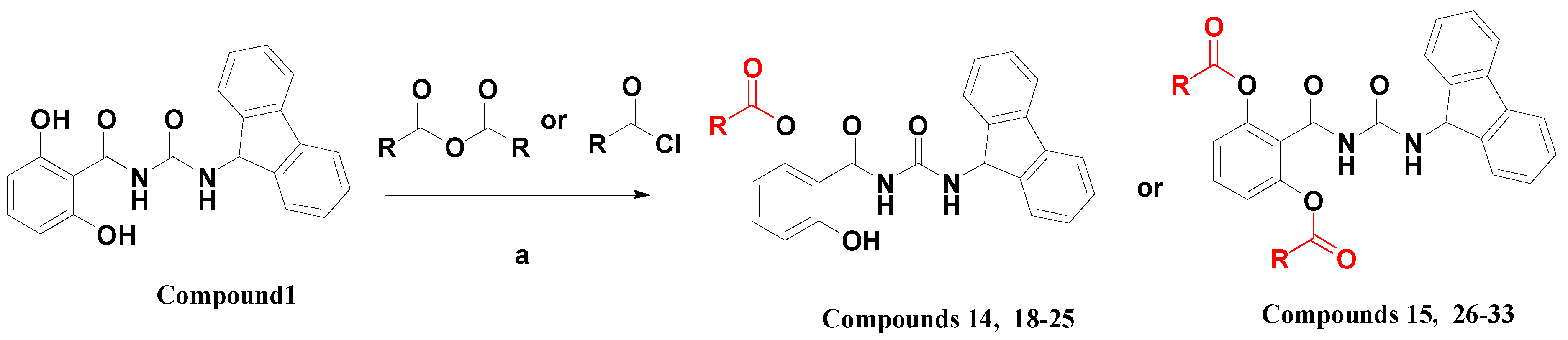
1-(2-Acetoxy-6-hydroxybenzoyl)-3-(9H-fluoren-9-yl)-urea (14). To a solution of 1 (0.9 g) and triethylamine (0.35 mL) in anhydrous DMF (25 mL), was added acetic anhydride (0.21 mL) with stirring at −20 °C. The mixture was stirred at room temperature for 12 h, and then heated to 40 °C with stirring for another 24 h. The reaction mixture was poured into H2O (500 mL), extracted with dichloromethane (DCM, 200 mL) three times. The organic layers were combined, dried with anhydrous sodium sulfate, and concentrated to dryness. The residue was purified by flash column chromatography on silica gel, eluted with a mixture of THF/DCM/petroleum ether (PE) (1:5:3, v/v/v), to afford 14 (161 mg, 16%) as a white solid: mp 215–225 °C; 1H-NMR (DMSO-d6) δ: 10.84 (s, 1H), 10.45 (s, 1H), 8.69 (s, 1H), 7.89 (d, J = 7.5 Hz, 2H), 7.62 (t, J = 7.5 Hz, 2H), 7.46 (t, J = 7.4 Hz, 2H), 7.37 (t, J = 7.4 Hz, 2H), 7.27 (t, J = 8.2 Hz, 1H), 6.80 (d, J = 8.3 Hz, 1H), 6.63 (d, J = 8.0 Hz, 1H), 5.97 (d, J = 7.9 Hz, 1H), 2.13 (s, 3H); ESI-MS m/z 401.0 [M − H]−; HRMS (ESI) m/z calcd C23H18N2O5 [M − H]− 401.1137, found 401.1143.
1-(2-Benzoyloxy-6-hydroxybenzoyl)-3-(9H-fluoren-9-yl)-urea (18). Prepared in the same manner as described for 14, but the acetic anhydride was replaced by benzoyl chloride: mp 211–214 °C; 1H-NMR (DMSO-d6) δ: 11.07 (s, 1H), 10.45 (s, 1H), 8.61 (s, 1H), 8.01 (d, J = 7.4 Hz, 2H), 7.85 (t, J = 7.7 Hz, 2H), 7.78 (t, J = 7.4 Hz, 1H), 7.58 (t, J = 7.7 Hz, 2H), 7.40 (dd, J = 14.5, 7.0 Hz, 2H), 7.38–7.30 (m, 3H), 7.26 (t, J = 7.4 Hz, 2H), 6.84 (t, J = 8.9 Hz, 2H), 5.91 (d, J = 8.2 Hz, 1H); ESI-MS m/z 463.0 [M − H]−; HRMS (ESI) m/z calcd C28H20N2O5 [M − H]− 463.1294, found 463.1299.
1-(2-Hydroxy-6-propionyloxybenzoyl)-3-(9H-fluoren-9-yl)-urea (19). Prepared in the same manner as described for 14, but the acetic anhydride was replaced by propionic anhydride: mp 206–216 °C; 1H-NMR (acetone-d6) δ: 9.72 (s, 1H), 8.71 (d, J = 8.1 Hz, 1H), 7.86 (d, J = 7.5 Hz, 2H), 7.70 (d, J = 7.4 Hz, 2H), 7.46 (t, J = 7.4 Hz, 2H), 7.43–7.34 (m, 3H), 6.89 (d, J = 8.4 Hz, 1H), 6.75 (d, J = 8.1 Hz, 1H), 6.11 (d, J = 8.1 Hz, 1H), 2.60 (q, J = 7.5 Hz, 2H), 1.23–1.12 (m, 3H); ESI-MS m/z 415.0 [M − H]−; HRMS (ESI) m/z calcd C24H20N2O5 [M − H]− 415.1294, found 415.1299.
1-(2-Hydroxy-6-n-butyryloxybenzoyl)-3-(9H-fluoren-9-yl)-urea (20). Prepared in the same manner as described for 14, but the acetic anhydride was replaced by butyric anhydride: mp 204–210 °C; 1H-NMR (acetone-d6) δ: 8.71 (s, 1H), 7.87 (d, J = 7.5 Hz, 2H), 7.71 (d, J = 7.5 Hz, 2H), 7.47 (t, J = 7.4 Hz, 2H), 7.44–7.36 (m, 3H), 6.91 (d, J = 7.7 Hz, 1H), 6.75 (d, J = 7.9 Hz, 1H), 6.12 (d, J = 8.2 Hz, 1H), 2.56 (t, J = 7.3 Hz, 2H), 1.76–1.64 (m, 2H), 0.96 (t, J = 7.4 Hz, 3H); ESI-MS m/z 429.1 [M − H]−; HRMS (ESI) m/z calcd C25H22N2O5 [M − H]− 429.1450, found 429.1456.
1-(2-Cyclopropyl formyloxy-6-hydroxybenzoyl)-3-(9H-fluoren-9-yl)-urea (21). Prepared in the same manner as described for 14, but the acetic anhydride was replaced by cyclopropanecarboxylic acid chloride: mp 194–199 °C; 1H-NMR (DMSO-d6) δ: 10.85 (s, 1H), 10.36 (s, 1H), 8.71 (s, 1H), 7.90 (d, J = 7.6 Hz, 2H), 7.61 (d, J = 7.5 Hz, 2H), 7.47 (t, J = 7.4 Hz, 2H), 7.38 (t, J = 7.5 Hz, 2H), 7.25 (t, J = 8.2 Hz, 1H), 6.79 (d, J = 8.2 Hz, 1H), 6.63 (d, J = 8.2 Hz, 1H), 6.00 (d, J = 8.1 Hz, 1H), 1.75 (d, J = 4.8 Hz, 1H), 0.99–0.83 (m, 4H); EI-MS m/z 428.1(M+); 165.1 (100%); HRMS (EI) m/z calcd C25H20 N2O5 (M+) 428.1372, found 428.1373.
1-(2-Hydroxy-6-valeryloxybenzoyl)-3-(9H-fluoren-9-yl)-urea (22). Prepared in the same manner as described for 14, but acetic anhydride was replaced by valeric anhydride: mp 193–199 °C; 1H-NMR (acetone-d6) δ: 8.73 (d, J = 6.2 Hz, 1H), 7.86 (d, J = 7.6 Hz, 2H), 7.70 (d, J = 7.5 Hz, 2H), 7.46 (t, J = 7.6 Hz, 2H), 7.43–7.35 (m, 3H), 6.89 (d, J = 8.3 Hz, 1H), 6.73 (d, J = 8.1 Hz, 1H), 6.11 (d, J = 8.1 Hz, 1H), 2.57 (t, J = 7.4 Hz, 2H), 1.64 (dt, J = 15.2, 7.5 Hz, 2H), 1.37 (dq, J = 14.7, 7.4 Hz, 2H), 0.86 (t, J = 7.4 Hz, 3H); ESI-MS m/z 443.0 [M − H]−; HRMS (ESI) m/z calcd C26H24N2O5 [M − H]− 443.1607, found 443.1612.
1-(2-Hydroxy-6-trimethylacetoxyloxybenzoyl)-3-(9H-fluoren-9-yl)-urea (23). Prepared in the same manner as described for 14, but the acetic anhydride was replaced by trimethylacetic anhydride: mp 187–189 °C; 1H-NMR (acetone-d6) δ: 9.71 (s, 1H), 8.76 (s, 1H), 7.86 (d, J = 7.5 Hz, 2H), 7.68 (d, J = 7.4 Hz, 2H), 7.46 (t, J = 7.4 Hz, 2H), 7.37 (dd, J = 13.2, 7.3 Hz, 3H), 6.87 (d, J = 8.2 Hz, 1H), 6.69 (d, J = 8.1 Hz, 1H), 6.11 (d, J = 7.9 Hz, 1H), 1.31 (s, 9H); ESI-MS m/z 443.0 [M − H]−; HRMS (ESI) m/z calcd C26H24N2O5 [M − H]− 443.1607, found 443.1612.
1-(2-Hexanoyloxy-6-hydroxybenzoyl)-3-(9H-fluoren-9-yl)-urea (24). Prepared in the same manner as described for 14, but the acetic anhydride was replaced by hexanoic anhydride: mp 200–222 °C; 1H-NMR (DMSO-d6) δ: 11.09 (s, 1H), 10.42 (s, 1H), 8.73 (s, 1H), 7.89 (d, J = 7.5 Hz, 2H), 7.60 (d, J = 7.4 Hz, 2H), 7.46 (t, J = 7.3 Hz, 2H), 7.36 (t, J = 7.3 Hz, 2H), 7.25 (t, J = 8.2 Hz, 1H), 6.77 (d, J = 8.3 Hz, 1H), 6.57 (d, J = 8.0 Hz, 1H), 5.98 (d, J = 7.8 Hz, 1H), 2.39 (t, J = 7.4 Hz, 2H), 1.48 (dd, J = 14.6, 7.3 Hz, 2H), 1.22–1.03 (m, 4H), 0.72 (t, J = 7.0 Hz, 3H). ; ESI-MS m/z 457.0 [M − H]−; HRMS (ESI) m/z calcd C27H26N2O5 [M − H]− 457.1763, found 457.1769.
1-(2-Hydroxy-6-n-Octanoyloxybenzoyl)-3-(9H-fluoren-9-yl)-urea (25). Prepared in the same manner as described for 14, but the acetic anhydride was replaced by n-octanoic anhydride: mp 184–189 °C; 1H-NMR (CDCl3) δ: 11.56 (s, 1H), 9.45 (s, 1H), 8.48 (d, J = 8.4 Hz, 1H), 7.73 (d, J = 7.6 Hz, 2H), 7.65 (d, J = 7.5 Hz, 2H), 7.43 (dd, J = 12.1, 4.8 Hz, 3H), 7.34 (t, J = 7.5 Hz, 2H), 6.89 (d, J = 8.4 Hz, 1H), 6.69 (d, J = 8.1 Hz, 1H), 6.16 (d, J = 8.4 Hz, 1H), 2.81 (t, J = 7.4 Hz, 2H), 1.85 (dt, J = 15.1, 7.4 Hz, 2H), 1.53–1.28 (m, 8H), 0.91 (t, J = 6.8 Hz, 3H); EI-MS m/z 486.2 (M+); 165.1 (100%); HRMS (EI) m/z calcd C29H30N2O5 (M+) 486.2155, found 486.2157.
1-[2,6-Bis-(acetoxy)benzoyl]-3-(9H-fluoren-9-yl)-urea (15). To a solution of 1 (0.9 g) and triethylamine (1.38 mL) in anhydrous DMF (15 mL), was added acetic anhydride (0.93 mL) with stirring at room temperature for 12 h. The reaction mixture was poured into H2O (300 mL), and extracted with DCM (120 mL) three times. The organic layers were combined, dried with anhydrous sodium sulfate, then concentrated to dryness. The residue was purified by flash column chromatography on silica gel, eluted with a mixture of THF/DCM/PE (1:5:3, v/v/v), to afford 15 (760 mg, 69%) as a white solid: mp 191–194 °C; 1H-NMR (acetone-d6) δ: 8.64 (s, 1H), 7.86 (d, J = 7.5 Hz, 2H), 7.69 (d, J = 7.5 Hz, 2H), 7.56 (t, J = 8.3 Hz, 1H), 7.46 (t, J = 7.4 Hz, 2H), 7.38 (t, J = 7.2 Hz, 2H), 7.18 (t, J = 8.7 Hz, 2H), 6.10 (d, J = 8.1 Hz, 1H), 2.24 (s, 6H); ESI-MS m/z 445.0 [M + H]+; HRMS (ESI) m/z calcd C25H20N2O6 [M + Na]+ 467.1219, found 467.1214.
1-[2,6-Bis-(benzoyloxy)benzoyl]-3-(9H-fluoren-9-yl)-urea (26). Prepared in the same manner as described for 15, but the acetic anhydride was replaced by benzoyl chloride: mp 211–213 °C; 1H-NMR (acetone-d6) δ: 10.33 (s, 1H), 8.54 (d, J = 7.0 Hz, 1H), 8.18 (d, J = 8.2 Hz, 4H), 7.84–7.68 (m, 5H), 7.62 (t, J = 7.7 Hz, 4H), 7.48 (d, J = 8.3 Hz, 2H), 7.37 (t, J = 7.5 Hz, 2H), 7.31 (d, J = 7.4 Hz, 2H), 7.21 (t, J = 7.4 Hz, 2H), 5.96 (d, J = 8.3 Hz, 1H); EI-MS m/z 568.2(M+); 105.0 (100%); HRMS (ESI) m/z calcd C35H24N2O6 [M + Na]+ 591.1532, found 591.1523.
1-[2,6-Bis-(propionyloxy)benzoyl]-3-(9H-fluoren-9-yl)-urea (27). Prepared in the same manner as described for 15, but the acetic anhydride was replaced by propionic anhydride: mp 165–167 °C; 1H-NMR (acetone-d6) δ: 9.89 (s, 1H), 8.67 (s, 1H), 7.87 (d, J = 7.6 Hz, 2H), 7.68 (d, J = 7.5 Hz, 2H), 7.58 (t, J = 8.3 Hz, 1H), 7.48 (t, J = 7.3 Hz, 2H), 7.43–7.35 (m, 2H), 7.20 (d, J = 8.3 Hz, 2H), 6.11 (d, J = 8.1 Hz, 1H), 2.59 (q, J = 7.5 Hz, 4H), 1.18 (t, J = 7.5 Hz, 6H); EI-MS m/z 472.2(M+); 165.1 (100%); HRMS (ESI) m/z calcd C27H24N2O6 [M + Na]+ 495.1532, found 495.1527.
1-[2,6-Bis-(n-butyryloxy)benzoyl]-3-(9H-fluoren-9-yl)-urea (28). Prepared in the same manner as described for 15, but the acetic anhydride was replaced by butyric anhydride: mp 186–188 °C; 1H-NMR (ad6) δ: 9.90 (s, 1H), 8.68 (s, 1H), 7.86 (d, J = 7.5 Hz, 2H), 7.67 (d, J = 7.5 Hz, 2H), 7.56 (t, J = 8.3 Hz, 1H), 7.47 (t, J = 7.3 Hz, 2H), 7.38 (td, J = 7.5, 1.0 Hz, 2H), 7.18 (d, J = 8.3 Hz, 2H), 6.10 (d, J = 8.1 Hz, 1H), 2.53 (t, J = 7.3 Hz, 4H), 1.78–1.62 (m, 4H), 0.96 (t, J = 7.4 Hz, 6H); EI-MS m/z 500.2 (M+); 165.1 (100%); HRMS (ESI) m/z calcd C29H28N2O6 [M + Na]+ 523.1845, found 523.1840.
1-[2,6-Bis-(cyclopropylformyloxy)benzoyl]-3-(9H-fluoren-9-yl)-urea (29). Prepared in the same manner as described for 15, but the acetic anhydride was replaced by cyclopropanecarboxylic acid chloride: mp 183–186 °C; 1H-NMR (CDCl3) δ: 8.50 (d, J = 8.0 Hz, 1H), 7.92 (s, 1H), 7.73 (d, J = 7.5 Hz, 2H), 7.65 (d, J = 7.5 Hz, 2H), 7.49 (t, J = 8.2 Hz, 1H), 7.43 (t, J = 7.4 Hz, 2H), 7.33 (t, J = 7.5 Hz, 2H), 7.08 (d, J = 8.2 Hz, 2H), 6.17 (d, J = 8.7 Hz, 1H), 1.84 (td, J = 7.8, 4.1 Hz, 2H), 1.21–1.13 (m, 4H), 1.08–0.99 (m, 4H); EI-MS m/z 496.2(M+); 165.1 (100%); HRMS (ESI) m/z calcd C29H24N2O6 [M + Na]+519.1532, found 519.1523.
1-[2,6-Bis-(valeryloxy)benzoyl]-3-(9H-fluoren-9-yl)-urea (30). Prepared in the same manner as described for 15, but the acetic anhydride was replaced by valeric anhydride: mp 172–174 °C; 1H-NMR (CDCl3) δ: 8.47 (d, J = 8.3 Hz, 1H), 8.01 (s, 1H), 7.72 (d, J = 7.5 Hz, 2H), 7.62 (d, J = 7.5 Hz, 2H), 7.49 (t, J = 8.2 Hz, 1H), 7.42 (t, J = 7.5 Hz, 2H), 7.31 (t, J = 7.4 Hz, 2H), 7.07 (d, J = 8.2 Hz, 2H), 6.13 (d, J = 8.4 Hz, 1H), 2.55 (t, J = 7.5 Hz, 4H), 1.75–1.63 (m, 4H), 1.38 (tt, J = 11.3, 5.8 Hz, 4H), 0.91 (t, J = 7.3 Hz, 6H); EI-MS m/z 528.2(M+); 165.1 (100%); HRMS (ESI) m/z calcd C31H32N2O6 [M + Na]+ 551.2158, found 551.2153.
1-[2,6-Bis-(trimethylacetoxyloxy)benzoyl]-3-(9H-fluoren-9-yl)-urea (31). Prepared in the same manner as described for 15, but the acetic anhydride was replaced by trimethylacetic anhydride: mp 202–205 °C; 1H-NMR (CDCl3) δ: 8.55 (d, J = 8.9 Hz, 1H), 7.71 (d, J = 7.5 Hz, 2H), 7.50 (t, J = 7.6 Hz, 2H), 7.49–7.37 (m, 3H), 7.28 (dd, J = 14.4, 6.9 Hz, 2H), 7.04 (d, J = 8.3 Hz, 2H), 6.13 (d, J = 8.7 Hz, 1H), 1.32 (s, 18H); EI-MS m/z 528.2(M+); 165.1 (100%); HRMS (ESI) m/z calcd C31H32N2O6 [M + Na]+ 551.2158, found 551.2153.
1-[2,6-Bis-(hexanoyloxy)benzoyl]-3-(9H-fluoren-9-yl)-urea (32). Prepared in the same manner as described for 15, but the acetic anhydride was replaced by hexanoic anhydride: mp 162–165 °C; 1H-NMR (CDCl3) δ: 8.46 (d, J = 8.0 Hz, 1H), 7.93 (s, 1H), 7.72 (d, J = 7.6 Hz, 2H), 7.62 (d, J = 7.4 Hz, 2H), 7.50 (t, J = 8.2 Hz, 1H), 7.42 (t, J = 7.4 Hz, 2H), 7.31 (dd, J = 14.1, 6.7 Hz, 2H), 7.07 (d, J = 8.3 Hz, 2H), 6.14 (d, J = 8.2 Hz, 1H), 2.54 (t, J = 7.6 Hz, 4H), 1.69 (dd, J = 14.6, 7.2 Hz, 4H), 1.30 (dd, J = 20.4, 5.5 Hz, 8H), 0.88 (t, J = 6.8 Hz, 6H); EI-MS m/z 556.3(M+); 165.1 (100%); HRMS (ESI) m/z calcd C33H36N2O6 [M + Na]+ 579.2471, found 579.2466.
1-[2,6-Bis-(n-Octanoyloxy)benzoyl]-3-(9H-fluoren-9-yl)-urea (33). Prepared in the same manner as described for 15, but the acetic anhydride was replaced by n-octanoic anhydride: mp 152–153 °C; 1H-NMR (CDCl3) δ: 8.48 (d, J = 8.3 Hz, 1H), 8.17 (s, 1H), 7.71 (t, J = 8.3 Hz, 2H), 7.61 (d, J = 7.5 Hz, 2H), 7.51–7.45 (m, 1H), 7.42 (t, J = 7.5 Hz, 2H), 7.31 (t, J = 7.5 Hz, 2H), 7.07 (d, J = 8.3 Hz, 2H), 6.12 (d, J = 8.4 Hz, 1H), 2.54 (t, J = 7.6 Hz, 4H), 1.77–1.63 (m, 4H), 1.34–1.17 (m, 16H), 0.87 (t, J = 6.3 Hz, 6H); HRMS (ESI) m/z calcd C37H44N2O6 [M + H]+ 613.3278, found 613.3267.