Two Methods for Increased Specificity and Sensitivity in Loop-Mediated Isothermal Amplification
Abstract
:1. Introduction
2. Results and Discussion
2.1. Non-Specific Amplification of LAMP Primers Targeting hlyA of L. Monocytogenes
| Results of Amplification | hlyA-FIP + hlyA-LF | hlyA-FIP + hlyA-LB | hlyA-FIP + hlyA-F3 | hlyA-FIP + hlyA-B3 | hlyA-BIP + hlyA-F3 | hlyA-BIP + hlyA-LF | hlyA-F3 + hlyA-B3 |
|---|---|---|---|---|---|---|---|
| Non-specific Amplification | 4/4 | 4/4 | 1/4 | 4/4 | 0/4 | 0/4 | 0/4 |
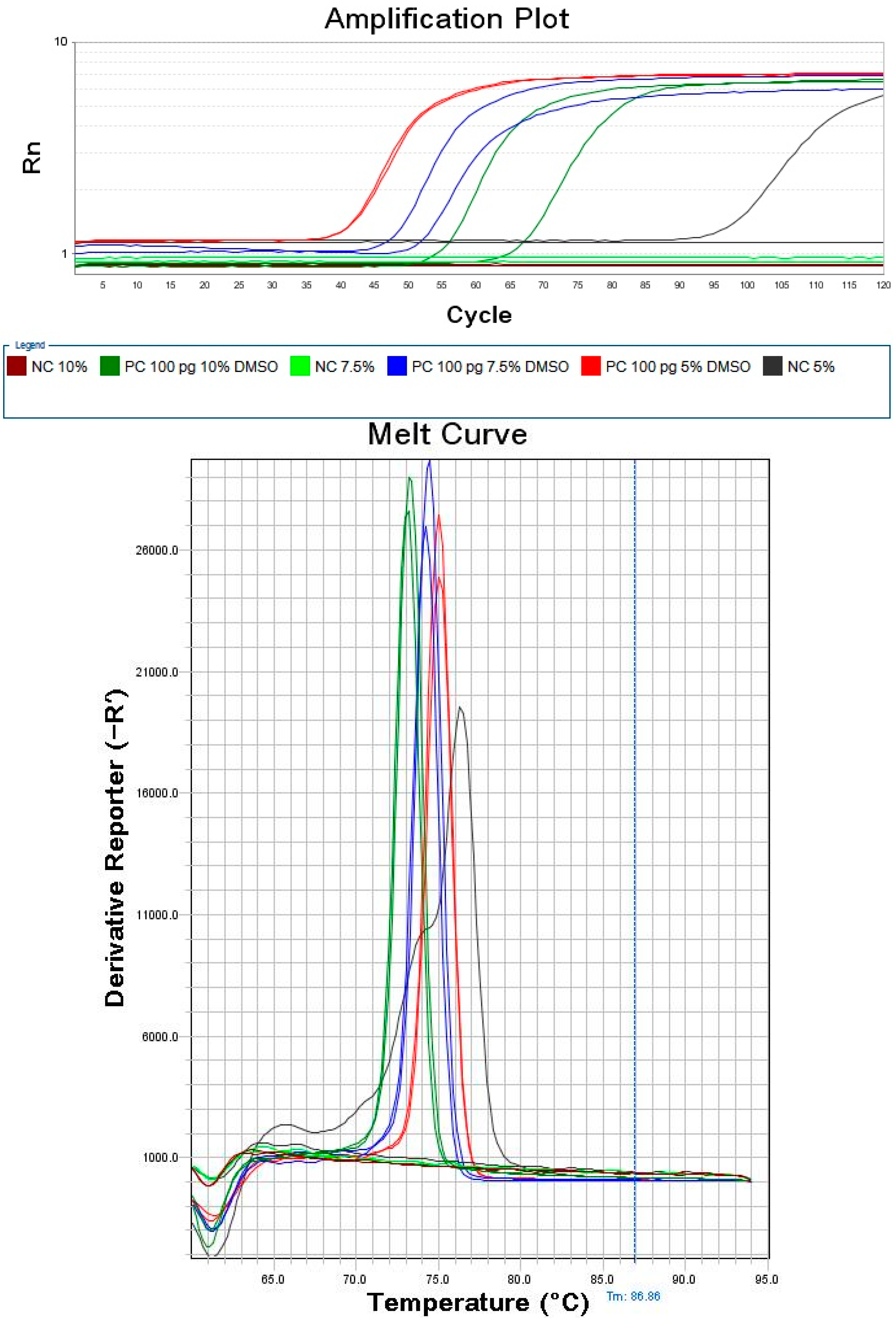
2.2. Optimization of DMSO Concentration
2.3. Selection of Reaction Temperature for LAMP
| PC/NC | Threshold Time at 61 °C (min) | Threshold Time at 59 °C (min) | Threshold Time at 57 °C (min) | Threshold Time at 55 °C (min) | Threshold Time at 53 °C (min) |
|---|---|---|---|---|---|
| Positive Control (1 pg DNA) | 42 | 28 | 26 | 28 | 36 |
| undetermined | 32 | 26 | 27 | 56 | |
| Negative Control | undetermined | undetermined | undetermined | undetermined | undetermined |
| undetermined | undetermined | undetermined | undetermined | undetermined |
2.4. Detection Limit Comparison
| Results of Amplification | Developed Conventional LAMP Assay 1 | Developed Touchdown LAMP Assay 1 | Reported LAMP Assay 2 | Isothermal Master Mix 3 | Loopamp®Listeria monocytogenes Detection Kit 4 |
|---|---|---|---|---|---|
| Detection Limit | 1000 fg (3/3) | 10 fg (2/3) | 1000 fg (1/3) | 100 fg (1/3) | 100 fg (3/3) |
| Non-specific Amplification of Negative Control | 0/5 | 0/5 | 1/4 | 0/5 | 0/5 |
2.5. Assaying Selectivity
| Bacterial Strain (Serotype) | Developed Touchdown LAMP Assay 1 | Reported LAMP Assay 2 | Isothermal Master Mix 3 | Loopamp®Listeria monocytogenes Detection Kit 4 |
|---|---|---|---|---|
| Listeria monocytogenes J1-225 (4b) | 3/3 | 2/2 | 3/3 | 3/3 |
| Listeria monocytogenes J2-020 (1/2a) | 3/3 | 2/2 | 3/3 | 3/3 |
| Listeria monocytogenes J2-064 (1/2b) | 3/3 | 1/2 | 3/3 | 3/3 |
| Listeria monocytogenes J1-169 (3b) | 3/3 | 2/2 | 3/3 | 3/3 |
| Listeria monocytogenes J1-049 (3c) | 3/3 | 2/2 | 3/3 | 3/3 |
| Listeria monocytogenes M1-004 (N/A) | 3/3 | 2/2 | 3/3 | 3/3 |
| Listeria monocytogenes J1-094 (1/2c) | 3/3 | 2/2 | 3/3 | 3/3 |
| Listeria monocytogenes C1-115 (3a) | 3/3 | 2/2 | 3/3 | 3/3 |
| Listeria monocytogenes J1-031 (4a) | 3/3 | 2/2 | 5/7 | 3/3 |
| Listeria monocytogenes W1-110 (4c) | 3/3 | 2/2 | 3/3 | 3/3 |
| Listeria monocytogenes ATCC19115 (4b) | 3/3 | 1/2 | 3/3 | 3/3 |
| Listeria innocua ATCC51742 | 0/3 | 0/2 | 0/3 | 1/7 |
| Listeria invanovii ATCC49954 | 1/7 | 0/2 | 0/3 | 0/3 |
| Salmonella typhimuriam | 0/3 | 0/2 | 0/3 | 0/3 |
| Salmonella enterica serotype Newport | 0/3 | 0/2 | 0/3 | 0/3 |
| Escherichia coli O157:H7 933 | 0/3 | 0/2 | 0/3 | 0/3 |
| Negative Controls | 0/7 | 0/3 | 1/6 | 1/8 |
3. Experimental Section
3.1. Primer Design
| Primer | Sequence (5'–3') |
|---|---|
| hlyA-FIP | CGTGTTTCTTTTCGATTGGCGTCTTTTTTTCATCCATGGCACCACC |
| hlyA-BIP | CCACGGAGATGCAGTGACAAATGTTTTGGATTTCTTCTTTTTCTCCACAAC |
| hlyA-F3 | TTGCGCAACAAACTGAAGC |
| hlyA-B3 | GCTTTTACGAGAGCACCTGG |
| hlyA-LF | TAGGACTTGCAGGCGGAGATG |
| hlyA-LB | GCCAAGAAAAGGTTACAAAGATGG |
| prfA-FIP | CCGCTCCTTTTTAATTCGTAAAACTTTTTAAAACGTGTCCTTAACTCTCTC |
| prfA-BIP | ATCATGGTAATAGCTTTTCAGGCTTTTTTTTGAAGTTTTTCTTCCCCG |
| prfA-F3 | AACGTATAATTTAGTTCCCACAT |
| prfA-B3 | GGGTCTTTTTGGCTTGTGTA |
| prfA-LF | TTAAGCCACCTACAACTAATCTGAC |
| prfA-LB | CATTTCACTATGACGGTAAAAGCAG |
3.2. Non-Specific Amplification of LAMP Primers
3.3. Optimization of DMSO Concentration
3.4. Selection of Reaction Temperature for LAMP
3.5. Sensitivity Comparison of Our Developed LAMP Assay, Reported LAMP Assay and Commercial Isothermal Amplification Kit
3.6. Specificity Determination of Optimized Touchdown LAMP assay, Tang LAMP Assay and Commercial Isothermal Amplification Kit
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucl. Acid. Res. 2000, 28, e63. [Google Scholar] [CrossRef]
- Nagamine, K.; Hase, T.; Notomi, T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 2002, 16, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Saiki, R.K.; Scharf, S.; Faloona, F.; Mullis, K.B.; Horn, G.T.; Erlich, H.A.; Arnheim, N. Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 1985, 230, 1350–1354. [Google Scholar] [CrossRef] [PubMed]
- Saiki, R.K.; Gelfand, D.H.; Stoffel, S.; Scharf, S.J.; Higuchi, R.; Horn, G.T.; Mullis, K.B.; Erlich, H.A. Primer-Directed Enzymatic Amplification of DNA with a Thermostable DNA Polymerase. Science 1998, 239, 487–491. [Google Scholar] [CrossRef]
- Compton, J. Nucleic acid sequence-based amplification. Nature 1991, 350, 91–92. [Google Scholar] [CrossRef] [PubMed]
- Fahy, E.; Kwoh, D.Y.; Gingeras, T.R. Self-sustained sequence replication (3SR): An isothermal transcription-based amplification system alternative to PCR. Genome. Res. 1991, 1, 25–33. [Google Scholar] [CrossRef]
- Walker, G.T.; Fraiser, M.S.; Schram, J.L.; Little, M.C.; Nadeau, J.G.; Malinowski, D.P. Strand displacement amplification-an isothermal, in vitro DNA amplification technique. Nucl. Acid Res. 1992, 20, 1691–1696. [Google Scholar] [CrossRef]
- Lizardi, P.M.; Huang, X.; Zhu, Z.; Bray-Ward, P.; Thomas, D.C.; Ward, D.C. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat. Genet. 1998, 19, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Vincent, M.; Yan, X.; Kong, H. Helicase-dependent isothermal DNA amplification. EMBO Rep. 2004, 5, 795–800. [Google Scholar] [CrossRef] [PubMed]
- Fang, R.; Li, X.; Hu, L.; You, Q.; Li, J.; Wu, J.; Xu, P.; Zhong, H.; Luo, Y.; Mei, J.; Gao, Q. Cross-priming amplification for rapid detection of Mycobacterium tuberculosis in sputum specimens. J. Clin. Microbiol. 2009, 47, 845–847. [Google Scholar] [CrossRef] [PubMed]
- Woźniakowski, G.; Niczyporuk, J.S.; Samorek-Salamonowicz, E.; Gaweł, A. The development and evaluation of cross-priming amplification for the detection of avian reovirus. J. Appl. Microbiol. 2015, 118, 528–536. [Google Scholar] [CrossRef] [PubMed]
- Haridas, D.V.; Pillai, D.; Manojkumar, B.; Nair, C.M.; Sherief, P.M. Optimisation of reverse transcriptase loop-mediated isothermal amplification assay for rapid detection of Macrobrachium rosenbergii noda virus and extra small virus in Macrobrachium rosenbergii. J. Virol. Methods 2010, 167, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Perera, N.; Aonuma, H.; Yoshimura, A.; Teramoto, T.; Iseki, H.; Nelson, B.; Igarashi, I.; Yagi, T.; Fukumoto, S.; Kanuka, H. Rapid identification of virus-carrying mosquitoes using reverse transcription-loop-mediated isothermal amplification. J. Virol. Methods 2009, 156, 32–36. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.F.; Li, P.C. Development of reverse transcription loop-mediated isothermal amplification for rapid detection of porcine epidemic diarrhea virus. Virus Genes 2011, 42, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Woźniakowski, G.; Kozdruń, W.; Samorek-Salamonowicz, E. Loop-mediated isothermal amplification for the detection of goose circovirus. Virol. J. 2012, 9, 110. [Google Scholar] [CrossRef]
- Wassermann, M.; Mackenstedt, U.; Romig, T. A loop-mediated isothermal amplification (LAMP) method for the identification of species within the Echinococcus granulosus complex. Vet. Pathol. 2014, 200, 97–103. [Google Scholar]
- Prompamorn, P.; Sithigorngul, P.; Rukpratanporn, S.; Longyant, S.; Sridulyakul, P.; Chaivisuthangkura, P. The development of loop-mediated isothermal amplification combined with lateral flow dipstick for detection of Vibrio parahaemolyticus. Lett. Appl. Microbiol. 2011, 52, 344–351. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, T.C.; Ferrari, H.F.; Bregano, L.C.; Silva-Frade, C.; Rosa, A.C.; Andrade, A.L. Visual detection of turkey coronavirus RNA in tissues and feces by reverse-transcription loop-mediated isothermal amplification (RT-LAMP) with hydroxynaphthol blue dye. Mol. Cell. Probes 2010, 24, 415–417. [Google Scholar] [CrossRef] [PubMed]
- Nzelu, C.O.; Gomez, E.A.; Cáceres, A.G.; Sakurai, T.; Martini-Robles, L.; Uezato, H.; Mimori, T.; Katakura, K.; Hashiguchi, Y.; Kato, H. Development of a loop-mediated isothermal amplification method for rapid mass-screening of sand flies for Leishmania infection. Acta Trop. 2014, 132, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Zhi, X.; Deng, M.; Yang, H.; Gao, G.; Wang, K.; Fu, H.L.; Zhang, Y.X.; Chen, D.; Cui, D.X. A novel HBV genotypes detecting system combined with microfluidic chip, loop-mediated isothermal amplification and GMR sensors. Biosens. Bioelectron. 2014, 54, 372–377. [Google Scholar] [CrossRef] [PubMed]
- Tong, Q.B.; Chen, R.; Zhang, Y.; Yang, G.J.; Takashi, K.; Rieko, F.S.; Lou, D.; Yang, K.; Wen, L.Y.; Lu, S.H.; et al. A new surveillance and response tool: Risk map of infected Oncomelania hupensis detected by Loop-mediated isothermal amplification (LAMP) from pooled samples. Acta Trop. 2015, 141, 170–177. [Google Scholar]
- Tang, M.J.; Zhou, S.; Zhang, X.Y.; Pu, J.H.; Ge, Q.L.; Tang, X.J.; Gao, Y.S. Rapid and Sensitive Detection of Listeria monocytogenes by Loop-Mediated Isothermal Amplification. Curr. Microbiol. 2011, 63, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Li, S.L.; zhang, X.B.; Wang, D.G.; Kuang, Y.Y.; Xu, Y.G. Simple and Rapid Method for Detecting Foodborne Shigella by a Loop-mediated Isothermal Amplification. J. Rapid Methods Autom. Microbiol. 2009, 17, 465–475. [Google Scholar] [CrossRef]
- Higuchi, R.; Fockler, C.; Dollinger, G.; Watson, R. Kinetic PCR analysis: Real-time monitoring of DNA amplification reactions. Biotechnology 1993, 11, 1026–1030. [Google Scholar] [CrossRef] [PubMed]
- Frackman, S.; Kobs, G.; Simpson, D.; Storts, D. Betaine and DMSO: Enhancing Agents for PCR. Promega Notes 1998, 65, 27–29. [Google Scholar]
- Hung, T.; Mak, K.; Fong, K. A specificity enhancer for polymerase chain reaction. Nucl. Acid Res. 1990, 18, 4953. [Google Scholar] [CrossRef]
- Bachmann, B.; Lüke, W.; Hunsmann, G. Improvement of PCR amplified DNA sequencing with the aid of detergents. Nucl. Acid Res. 1990, 18, 1309. [Google Scholar] [CrossRef]
- Korbie, D.J.; Mattick, J.S. Touchdown PCR for increased specificity and sensitivity in PCR amplification. Nat. Protoc. 2008, 3, 1452–1456. [Google Scholar] [CrossRef] [PubMed]
- Don, R.H.; Cox, P.T.; Wainwright, B.J.; Baker, K.; Mattick, J.S. “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucl. Acid Res. 1991, 19, 4008. [Google Scholar] [CrossRef]
- Sample Availability: Not available.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.-G.; Brewster, J.D.; Paul, M.; Tomasula, P.M. Two Methods for Increased Specificity and Sensitivity in Loop-Mediated Isothermal Amplification. Molecules 2015, 20, 6048-6059. https://doi.org/10.3390/molecules20046048
Wang D-G, Brewster JD, Paul M, Tomasula PM. Two Methods for Increased Specificity and Sensitivity in Loop-Mediated Isothermal Amplification. Molecules. 2015; 20(4):6048-6059. https://doi.org/10.3390/molecules20046048
Chicago/Turabian StyleWang, De-Guo, Jeffrey D. Brewster, Moushumi Paul, and Peggy M. Tomasula. 2015. "Two Methods for Increased Specificity and Sensitivity in Loop-Mediated Isothermal Amplification" Molecules 20, no. 4: 6048-6059. https://doi.org/10.3390/molecules20046048






