Potential of Start Codon Targeted (SCoT) Markers to Estimate Genetic Diversity and Relationships among Chinese Elymus sibiricus Accessions
Abstract
:1. Introduction
2. Results and Discussion
2.1. Polymorphism of SCoT Markers
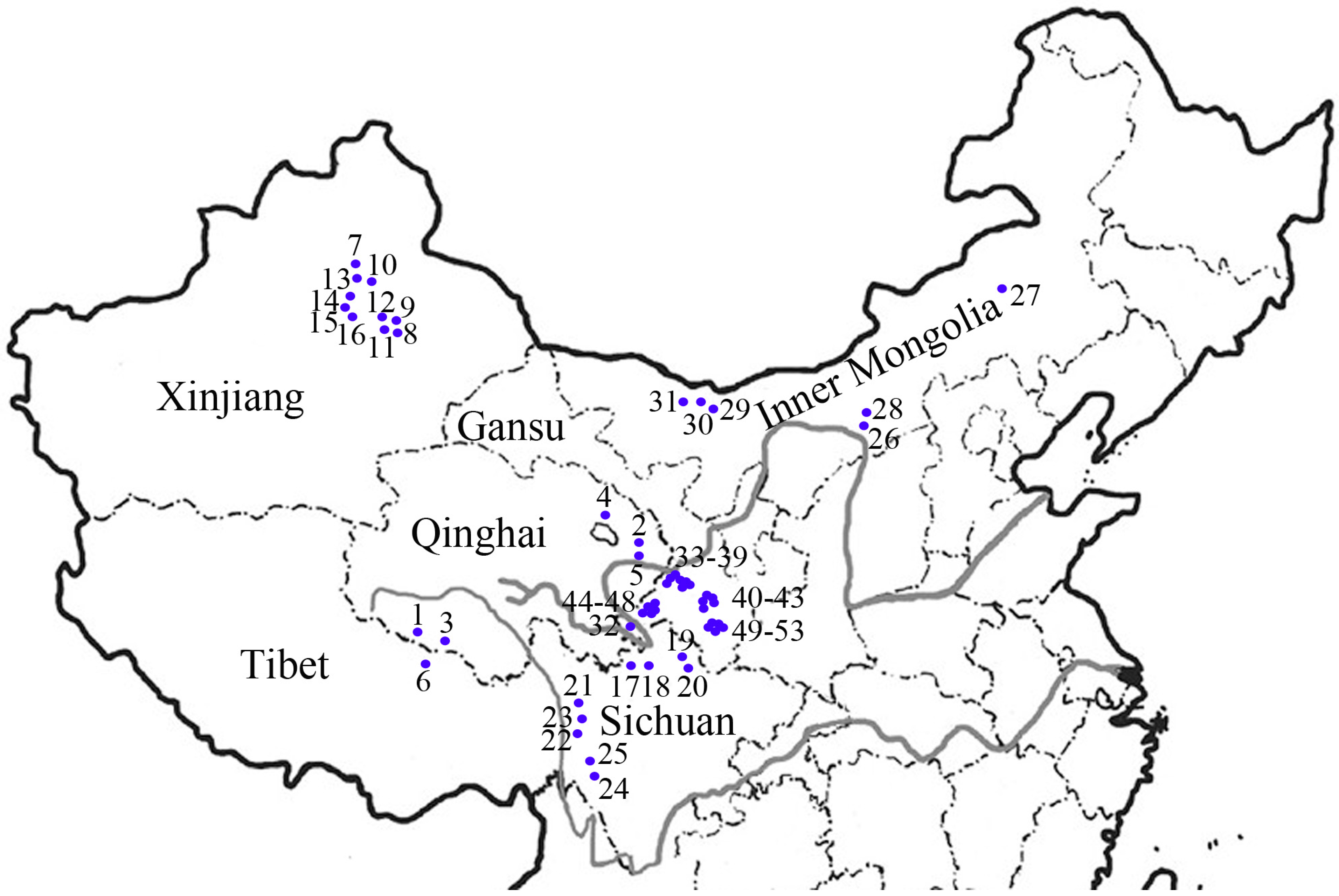
| POP | Code | Accession | Origin | Status |
|---|---|---|---|---|
| QX | 1 | Tongde | Qinghai, China | Cultivar |
| QX | 2 | PI504462 | Qinghai, China | Wild |
| QX | 3 | Qingmu1 | Qinghai, China | Cultivar |
| QX | 4 | PI504463 | Qinghai, China | Wild |
| QX | 5 | PI531669 | Qinghai, China | Wild |
| QX | 6 | PI639859 | Tibet, China | Wild |
| XJ | 7 | PI499468 | Xinjiang, China | Cultivated |
| XJ | 8 | PI499462 | Xinjiang, China | Wild |
| XJ | 9 | PI619577 | Xinjiang, China | Wild |
| XJ | 10 | PI595182 | Xinjiang, China | Wild |
| XJ | 11 | PI499614 | Xinjiang, China | Wild |
| XJ | 12 | PI499617 | Xinjiang, China | Wild |
| XJ | 13 | PI499619 | Xinjiang, China | Wild |
| XJ | 14 | PI595180 | Xinjiang, China | Wild |
| XJ | 15 | PI655140 | Xinjiang, China | Wild |
| XJ | 16 | Y2003 | Xinjiang, China | Wild |
| SC | 17 | Y1005 | Sichuan, China | Wild |
| SC | 18 | Chuancao2 | Hongyuan, Sichuan, China | Cultivar |
| SC | 19 | Hongyuan | Hongyuan, Sichuan, China | Breeding line |
| SC | 20 | SAU133 | Aba, Sichuan, China | wild |
| SC | 21 | SAU139 | Kangding, Sichuan, China | Wild |
| SC | 22 | SAU003 | Kangding, Sichuan, China | wild |
| SC | 23 | SAU137 | Aba, Sichuan, China | Wild |
| SC | 24 | SC02 | Ruoergai, Sichuan, China | Wild |
| SC | 25 | SC03 | Ruoergai, Sichuan, China | Wild |
| NM | 26 | PI499457 | Inner Mongolia, China | Cultivated |
| NM | 27 | PI499453 | Inner Mongolia, China | Wild |
| NM | 28 | PI499456 | Inner Mongolia, China | Cultivated |
| NM | 29 | PI499458 | Inner Mongolia, China | Cultivated |
| NM | 30 | PI499459 | Inner Mongolia, China | Cultivated |
| NM | 31 | W6 614214 | Inner Mongolia, China | Cultivated |
| GS | 32 | MQ01 | Maqu, Gansu, China | Wild |
| GS | 33 | HZ01 | Hezuo, Gansu, China | Wild |
| GS | 34 | HZ02 | Hezuo, Gansu, China | Wild |
| GS | 35 | HZ03 | Hezuo, Gansu, China | Wild |
| GS | 36 | XH03 | Xiahe, Gansu, China | Wild |
| GS | 37 | XH09 | Xiahe, Gansu, China | Wild |
| GS | 38 | XH02 | Xiahe, Gansu, China | Wild |
| GS | 39 | XH06 | Xiahe, Gansu, China | Wild |
| GS | 40 | LT04 | Lintan, Gansu, China | Wild |
| GS | 41 | LT02 | Lintan, Gansu, China | Wild |
| GS | 42 | LT05 | Lintan, Gansu, China | Wild |
| GS | 43 | LT01 | Lintan, Gansu, China | Wild |
| GS | 44 | LQ01 | Luqu, Gansu, China | Wild |
| GS | 45 | LQ03 | Luqu, Gansu, China | Wild |
| GS | 46 | LQ04 | Luqu, Gansu, China | Wild |
| GS | 47 | LQ09 | Luqu, Gansu, China | Wild |
| GS | 48 | LQ10 | Luqu, Gansu, China | Wild |
| GS | 49 | ZHN01 | Zhuoni, Gansu, China | Wild |
| GS | 50 | ZHN05 | Zhuoni, Gansu, China | Wild |
| GS | 51 | ZHN06 | Zhuoni, Gansu, China | Wild |
| GS | 52 | ZHN03 | Zhuoni, Gansu, China | Wild |
| GS | 53 | ZHN04 | Zhuoni, Gansu, China | Wild |
| Primer ID | Primer Sequence (5'–3') | TB | PB | PPB(%) | PIC | Rp |
|---|---|---|---|---|---|---|
| SCoT 5 | CAACAATGGCTACCACGA | 10 | 9 | 90.00 | 0.45 | 2.65 |
| SCoT 7 | CAACAATGGCTACCACGG | 7 | 6 | 85.7 | 0.41 | 2.53 |
| SCoT 9 | CAACAATGGCTACCAGCA | 12 | 9 | 75.00 | 0.44 | 3.35 |
| SCoT 10 | CAACAATGGCTACCAGCC | 13 | 11 | 84.62 | 0.45 | 2.40 |
| SCoT 23 | CACCATGGCTACCACCAG | 18 | 18 | 100.00 | 0.47 | 5.20 |
| SCoT 26 | ACCATGGCTACCACCGTC | 10 | 10 | 100.00 | 0.47 | 3.59 |
| SCoT 30 | CCATGGCTACCACCGGCG | 6 | 5 | 83.33 | 0.46 | 2.71 |
| SCoT 35 | CATGGCTACCACCGGCCC | 7 | 6 | 85.71 | 0.48 | 3.56 |
| SCoT 41 | CAATGGCTACCACTGACA | 17 | 16 | 94.12 | 0.50 | 6.49 |
| SCoT 42 | CAATGGCTACCATTAGCG | 11 | 10 | 90.91 | 0.50 | 2.56 |
| SCoT 44 | CAATGGCTACCATTAGCC | 7 | 7 | 100.00 | 0.47 | 1.94 |
| SCoT 45 | ACAATGGCTACCACTGAC | 11 | 7 | 63.64 | 0.45 | 2.14 |
| SCoT 54 | ACAATGGCTACCACCAGC | 6 | 4 | 66.67 | 0.20 | 1.40 |
| SCoT 60 | ACAATGGCTACCACCACA | 12 | 9 | 75.00 | 0.47 | 3.67 |
| SCoT 61 | CAACAATGGCTACCACCG | 10 | 9 | 90.00 | 0.46 | 3.32 |
| SCoT 62 | ACCATGGCTACCACGGAG | 16 | 14 | 87.50 | 0.50 | 3.86 |
| Mean | 10.8 | 9.4 | 89.60 | 0.45 | --- | |
| Total | 173 | 159 | 91.91 | --- | 51.37 |
2.2. Genetic Diversity Analysis
2.2.1. Genetic Diversity among Regions
| POP | NPB | PPB (%) | I | H | Na |
|---|---|---|---|---|---|
| QX | 97 | 56.07 | 0.3002 | 0.2217 | 1.5797 |
| XJ | 56 | 32.37 | 0.1662 | 0.1113 | 1.3237 |
| SC | 65 | 35.57 | 0.1978 | 0.1327 | 1.3757 |
| NM | 52 | 30.06 | 0.1657 | 0.1119 | 1.3006 |
| GS | 59 | 34.10 | 0.1615 | 0.1068 | 1.3410 |
| Mean | 65.8 | 37.63 | 0.1983 | 0.1369 | 1.3841 |
2.2.2. Genetic Diversity within Regions
| Source of Variance | d.f. | Sum of Squares | Variance Component | Total Variation (%) |
|---|---|---|---|---|
| Among geographic regions | 4 | 408.93 | 9.45 | 49.01 |
| Within geographic regions | 48 | 472.09 | 9.83 | 50.99 |
2.3. Population Structure and Cluster Analysis
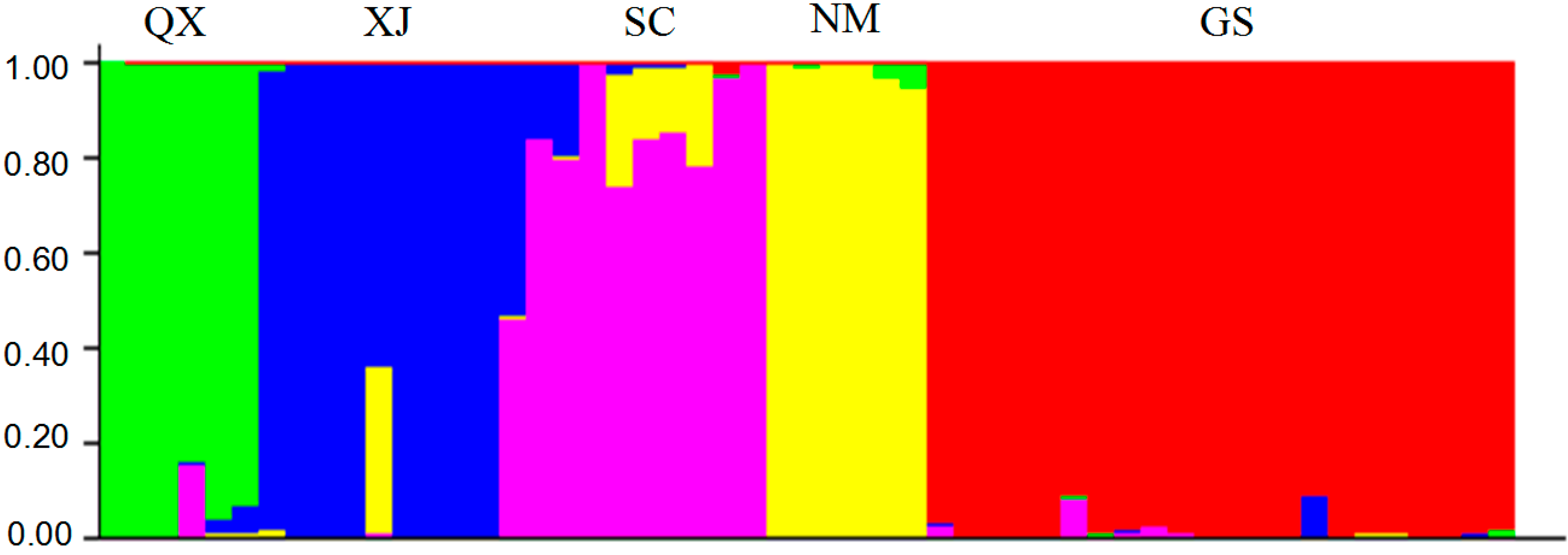
2.4. Conservation Implications
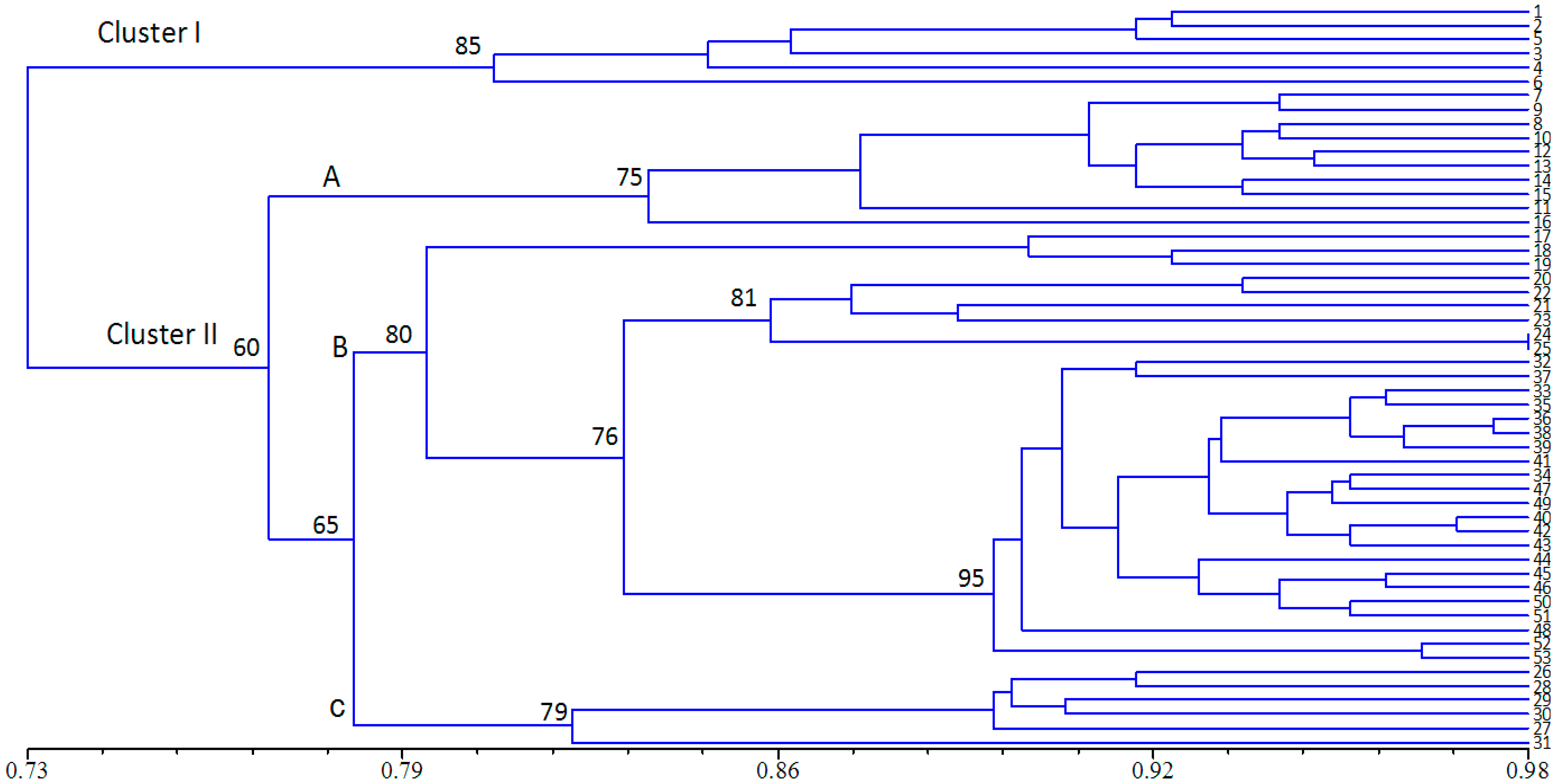
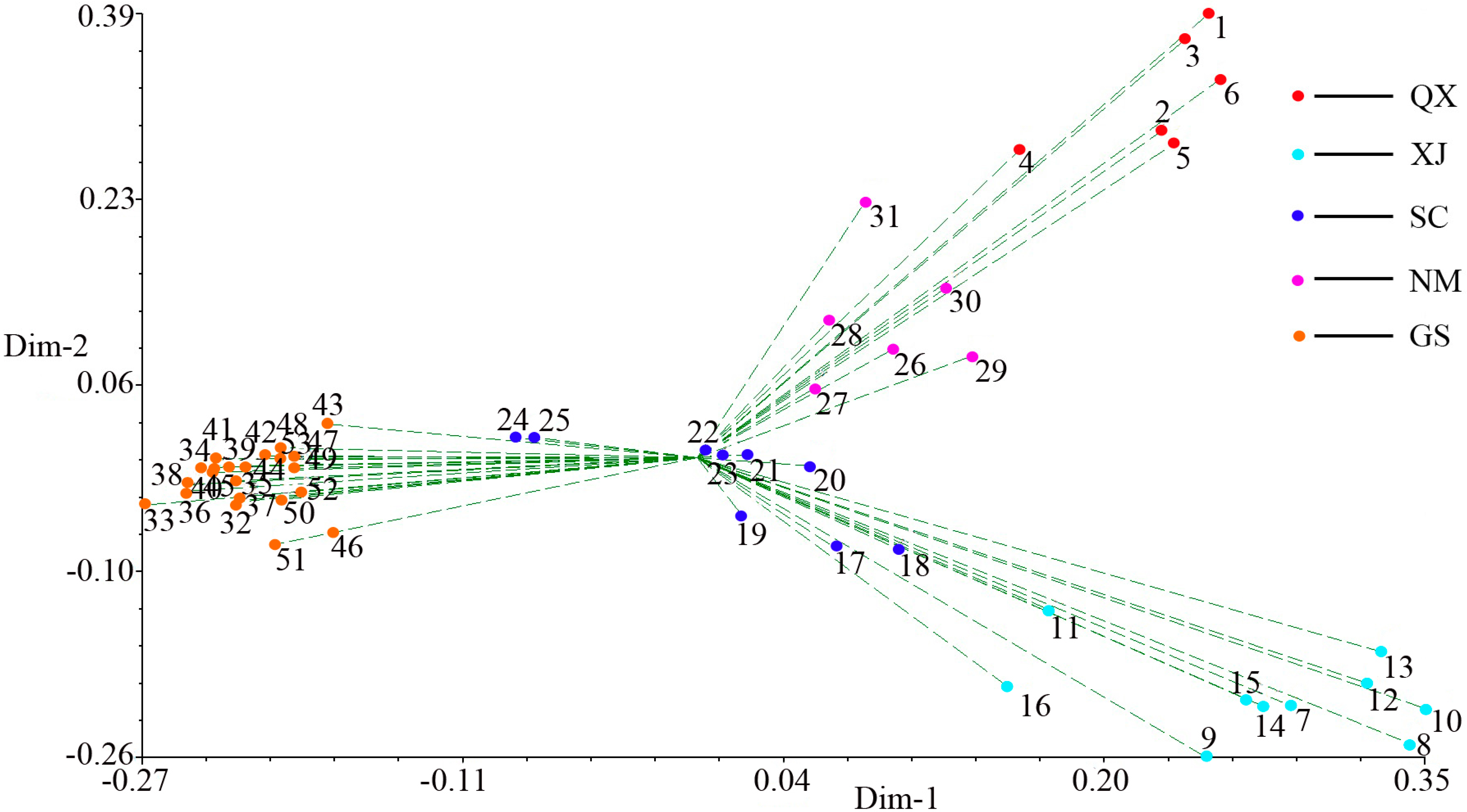
3. Experimental Section
3.1. Plant Materials
3.2. DNA Extraction
3.3. SCoT-PCR Amplification
3.4. Data Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Dewey, D.R. The Genomic system of classification as a guide to intergeneric hybridization with the perennial triticeae. In Gene Manipulation in Plant Improvement; Gustafson, J.P., Ed.; Plenum: New York, NY, USA, 1984; pp. 209–279. [Google Scholar]
- Bowden, W.M.; Cody, W.J. Recognition of Elymus sibiricus L. from Alaska and the district of Mackenzie. Bull. Torrey Bot. Club 1961, 88, 153–155. [Google Scholar] [CrossRef]
- Yan, J.J.; Bai, S.Q.; Ma, X.; Gan, Y.M.; Zhang, J.B. Genetic diversity of Elymus sibiricus and its breeding in China (In Chinese with English abstract). Chin. Bull. Bot. 2007, 24, 226–231. [Google Scholar]
- Futuyma, D.J. Evolutionary Biology; Sinauer Associates: Sunderland, MA, USA, 1986; p. 2. [Google Scholar]
- Ma, X.; Zhang, X.Q.; Zhou, Y.H.; Bai, S.Q.; Liu, W. Assessing genetic diversity of Elymus sibiricus (Poaceae: Triticeae) populations from Qinghai-Tibet Plateau by ISSR markers. Biochem. Syst. Ecol. 2008, 36, 514–522. [Google Scholar] [CrossRef]
- Yan, J.J.; Bai, S.Q.; Zhang, X.Q.; You, M.H.; Zhang, C.B.; Li, D.X.; Zeng, Y. Genetic diversity of wild Elymus sibiricus germplasm from the Qinghai-Tibetan Plateau in China detected by SRAP markers (In Chinese with English abstract). Acta Prataculturae Sin. 2010, 19, 173–183. [Google Scholar]
- Xie, W.G.; Zhao, X.H.; Zhang, J.Q.; Wang, Y.R.; Liu, W.X. Assessment of genetic diversity of Siberian wild rye (Elymus sibiricus L.) germplasms with variation of seed shattering and implication for future genetic improvement. Biochem. Syst. Ecol. 2015, 58, 211–218. [Google Scholar]
- Ma, X.; Chen, S.Y.; Zhang, X.Q.; Bai, S.Q.; Zhang, C.B. Assessment of Worldwide Genetic Diversity of Siberian Wild Rye (Elymus sibiricus L.) Germplasm Based on Gliadin Analysis. Molecules 2012, 17, 4424–4434. [Google Scholar]
- Collard, B.C.; Mackill, D.J. Start Codon Targeted (SCoT) polymorphism: A simple, novel DNA marker technique for generating gene-targeted markers in plants. Plan. Mol. Biol. Rep. 2009, 27, 86–93. [Google Scholar] [CrossRef]
- Mulpuri, S.; Muddanuru, T.; Francis, G. Start codon targeted (SCoT) polymorphism in toxic and non-toxic accessions of Jatropha curcas L. and development of a codominant SCAR marker. Plan. Sci. 2013, 207, 117–127. [Google Scholar]
- Cabo, S.; Ferreira, L.; Carvalho, A.; Martins-Lopes, P.; Martín, A.; Lima-Brito, J.E. Potential of Start Codon Targeted (SCoT) markers for DNA fingerprinting of newly synthesized tritordeums and their respective parents. J. Appl. Genet. 2014, 55, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Que, Y.X.; Pan, Y.B.; Lu, Y.H.; Yang, C.; Yang, Y.T.; Huang, N.; Xu, L.P. Genetic analysis of diversity within a Chinese local sugarcane germplasm based on start codon targeted polymorphism. BioMed Res. Int. 2014, 2014, 468375. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.L.; Zhang, J.Y.; Liu, C.H. Genetic diversity in some grape varieties revealed by SCoT analyses. Mol. Biol. Rep. 2012, 39, 5307–5313. [Google Scholar] [CrossRef] [PubMed]
- Gorji, A.M.; Matyas, K.K.; Dublecz, Z.; Decsi, K.; Cernak, I.; Hoffmann, B.; Taller, J.; Polgar, Z. In vitro osmotic stress tolerance in potato and identification of major QTLs. Am. J. Potato Res. 2012, 89, 453–464. [Google Scholar] [CrossRef]
- Luo, C.; He, X.H.; Chen, H.; Ou, S.J.; Gao, M.P. Analysis of diversity and relationships among mango cultivars using Start Codon Targeted (SCoT) markers. Biochem. Syst. Ecol. 2010, 38, 1176–1184. [Google Scholar] [CrossRef]
- Chen, F.Y.; Liu, J.H. Germplasm genetic diversity of Myrica rubra in Zhejiang Province studied using inter-primer binding site and start codon-targeted polymorphism markers. Sci. Hortic. 2014, 170, 169–175. [Google Scholar] [CrossRef]
- Gorji, A.M.; Poczai, P.; Polgar, Z.; Taller, J. Efficiency of arbitrarily amplified dominant markers (SCoT, ISSR and RAPD) for diagnostic fingerprinting in tetraploid potato. Am. J. Potato Res. 2011, 88, 226–237. [Google Scholar] [CrossRef]
- Xiong, F.Q.; Zhong, R.C.; Han, Z.Q.; Jiang, J.; He, L.Q.; Zhuang, W.J.; Tang, R.H. Start codon targeted polymorphism for evaluation of functional genetic variation and relationships in cultivated peanut (Arachis hypogaea L.) genotypes. Mol. Biol. Rep. 2011, 38, 3487–3494. [Google Scholar] [CrossRef]
- Amirmoradi, B.; Talebi, R.; Karami, E. Comparison of genetic variation and differentiation among annual Cicer species using start codon targeted (SCoT) polymorphism, DAMD-PCR, and ISSR markers. Plant Syst. Evol. 2012, 298, 1679–1688. [Google Scholar] [CrossRef]
- Yan, J.J.; Bai, S.Q.; Zhang, X.Q.; Chang, D.; You, M.H.; Zhang, C.B.; Li, D.X. Genetic diversity of native Elymus sibiricus populations in the Southeastern Margin of Qinghai-Tibetan Plateau as detected by SRAP and SSR markers (In Chinese with English abstract). Acta Prataculturae Sin. 2010, 19, 122–134. [Google Scholar]
- Ma, X.; Chen, S.Y.; Bai, S.Q.; Zhang, X.Q.; Li, D.X.; Zhang, C.B.; Yan, J.J. RAPD analysis of genetic diversity and population structure of Elymus sibiricus (Poaceae) native to the southeastern Qinghai-Tibet Plateau, China. Genet. Mol. Res. 2012, 11, 2708–2718. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.F.; Qi, X.; Zhang, X.Q.; Huang, L.K.; Ma, X.; Xie, W.G. Analysis of diversity and relationships among orchardgrass (Dactylis glomerata L.) accessions using start codon-targeted markers. Genet. Mol. Res. 2014, 13, 4406–4418. [Google Scholar]
- Xie, W.G.; Zhang, X.Q.; Ma, X.; Huang, L.K.; Zeng, B. Genetic variation of Dactylis glomerata germplasm from Southwest China detected by SSR markers (In Chinese with English abstract). Acta Pratac. Sin. 2009, 18, 138–146. [Google Scholar]
- Yan, X.B.; Guo, Y.X.; Zhou, H.; Wang, K. Analysis of geographical conditions affected on genetic variation and relationship among populations of Elymus (In Chinese with English abstract). J. Plan. Resour. Environ. 2006, 15, 17–24. [Google Scholar]
- Xie, W.G.; Zhang, X.Q.; Ma, X.; Cai, H.W.; Huang, L.K.; Peng, Y.; Zeng, B. Diversity comparison and phylogenetic relationships of cocksfoot (Dactylis glomerata L.) germplasm as revealed by SSR markers.) germplasm as revealed by SSR markers. Can. J. Plant. Sci. 2010, 90, 13–21. [Google Scholar]
- Wilson, B.L.; Kitzmiller, J.; Rolle, W.; Hipkins, V.D. Isozyme variation and its environmental correlates in Elymus glaucus from the California Floristic Province. Can. J. Bot. 2001, 79, 139–153. [Google Scholar]
- MacRitchie, D.; Sun, G. Evaluating the potential of barley and wheat microsatellite markers for genetic analysis of Elymus trachycaulus complex species. Theor. Appl. Genet. 2004, 108, 720–724. [Google Scholar] [CrossRef] [PubMed]
- Schoen, D.J.; Brown, A. Intraspecific variation in population gene diversity and effective population size correlates with the mating system in plants. Proc. Natl. Acad. Sci. USA 1991, 88, 4494–4497. [Google Scholar] [CrossRef] [PubMed]
- Hogbin, P.M.; Peakall, R. Evaluation of the contribution of genetic research to the management of the endangered plant Zieria prostrata. Conserv. Biol. 1999, 13, 514–522. [Google Scholar] [CrossRef]
- Ellstrand, N.C.; Elam, D.R. Population genetic consequences of small population size: Implications for plant conservation. Annu. Rev. Ecol. Syst. 1993, 24, 217–242. [Google Scholar] [CrossRef]
- Jin, Y.; Lu, B.R. Sampling strategy for genetic diversity (In Chinese with English abstract). Chin. Biodivers. 2002, 11, 155–161. [Google Scholar]
- Chen, S.Y.; Zhang, X.Q.; Ma, X.; Huang, L.K. Assessment of genetic diversity and differentiation of Elymus nutans indigenous to Qinghai-Tibet Plateau using simple sequence repeats markers. Can. J. Plant. Sci. 2013, 93, 1089–1096. [Google Scholar] [CrossRef]
- Jensen, K.B.; Mott, I.W.; Robins, J.G.; Waldron, B.L.; Nelson, M. Genetic improvement and diversity in snake River Wheatgrass (Elymus wawawaiensis) (Poaceae: Triticeae). Rangel. Ecol. Manag. 2012, 65, 76–84. [Google Scholar] [CrossRef]
- Robins, J.G.; Bushman, B.S.; Jensen, K.B. Dry matter yield combining ability among nine sources of cocksfoot germplasm. Euphytica 2012, 188, 419–428. [Google Scholar] [CrossRef]
- Ghislain, M.; Zhang, D.P.; Fajardo, D.; Huamán, Z.; Hijmans, R.J. Marker-assisted sampling of the cultivated Andean potato Solanum phureja collection using RAPD markers. Genet. Resour. Crop Evol. 1999, 46, 547–555. [Google Scholar] [CrossRef]
- Prevost, A.; Wilkinson, M. A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor. Appl. Genet. 1999, 98, 107–112. [Google Scholar] [CrossRef]
- Yeh, F.; Yang, R.; Boyle, T. POPGENE Version 1.31,Microsoft Windows-Based Freeware for Population Genetic Analysis: Quick User Guide; University of Alberta: Edmonton, AB, Canada, 1999. [Google Scholar]
- Mengoni, A.; Bazzicalupo, M. The statistical treatment of data and the analysis of molecular variance (AMOVA) in molecular microbial ecology. Ann. Microbiol. 2002, 52, 95–102. [Google Scholar]
- Rohlf, F.J. NTSYS-pc: Numerical Taxonomy and Multivariate Analysis System; Exeter Software: New York, NY, USA, 1992. [Google Scholar]
- Yap, I.V.; Nelson, R.J. Winboot: A Program for Performing Bootstrap Analysis of Binary Data to Determine the Confidence Limits of UPGMA-based Dendrograms; International Rice Research Institute: Manila, Philippines, 1996. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure from multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.M.; Ge, S. Data analysis in population genetics. I. analysis of RAPD data with AMOVA (In Chinese with English abstract). Chin. Biodivers. 2002, 4, 438–444. [Google Scholar]
- Poczai, P.; Varga, I.; Laos, M.; Cseh, A.; Bell, N.; Valkonen, J.P.T.; Hyvönen, J. Advances in plant gene-targeted and functional markers: A review. Plant Methods 2013, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds are available from the authors.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Xie, W.; Wang, Y.; Zhao, X. Potential of Start Codon Targeted (SCoT) Markers to Estimate Genetic Diversity and Relationships among Chinese Elymus sibiricus Accessions. Molecules 2015, 20, 5987-6001. https://doi.org/10.3390/molecules20045987
Zhang J, Xie W, Wang Y, Zhao X. Potential of Start Codon Targeted (SCoT) Markers to Estimate Genetic Diversity and Relationships among Chinese Elymus sibiricus Accessions. Molecules. 2015; 20(4):5987-6001. https://doi.org/10.3390/molecules20045987
Chicago/Turabian StyleZhang, Junchao, Wengang Xie, Yanrong Wang, and Xuhong Zhao. 2015. "Potential of Start Codon Targeted (SCoT) Markers to Estimate Genetic Diversity and Relationships among Chinese Elymus sibiricus Accessions" Molecules 20, no. 4: 5987-6001. https://doi.org/10.3390/molecules20045987





