Synthesis, Antifungal Activity and QSAR of Some Novel Carboxylic Acid Amides
Abstract
:1. Introduction
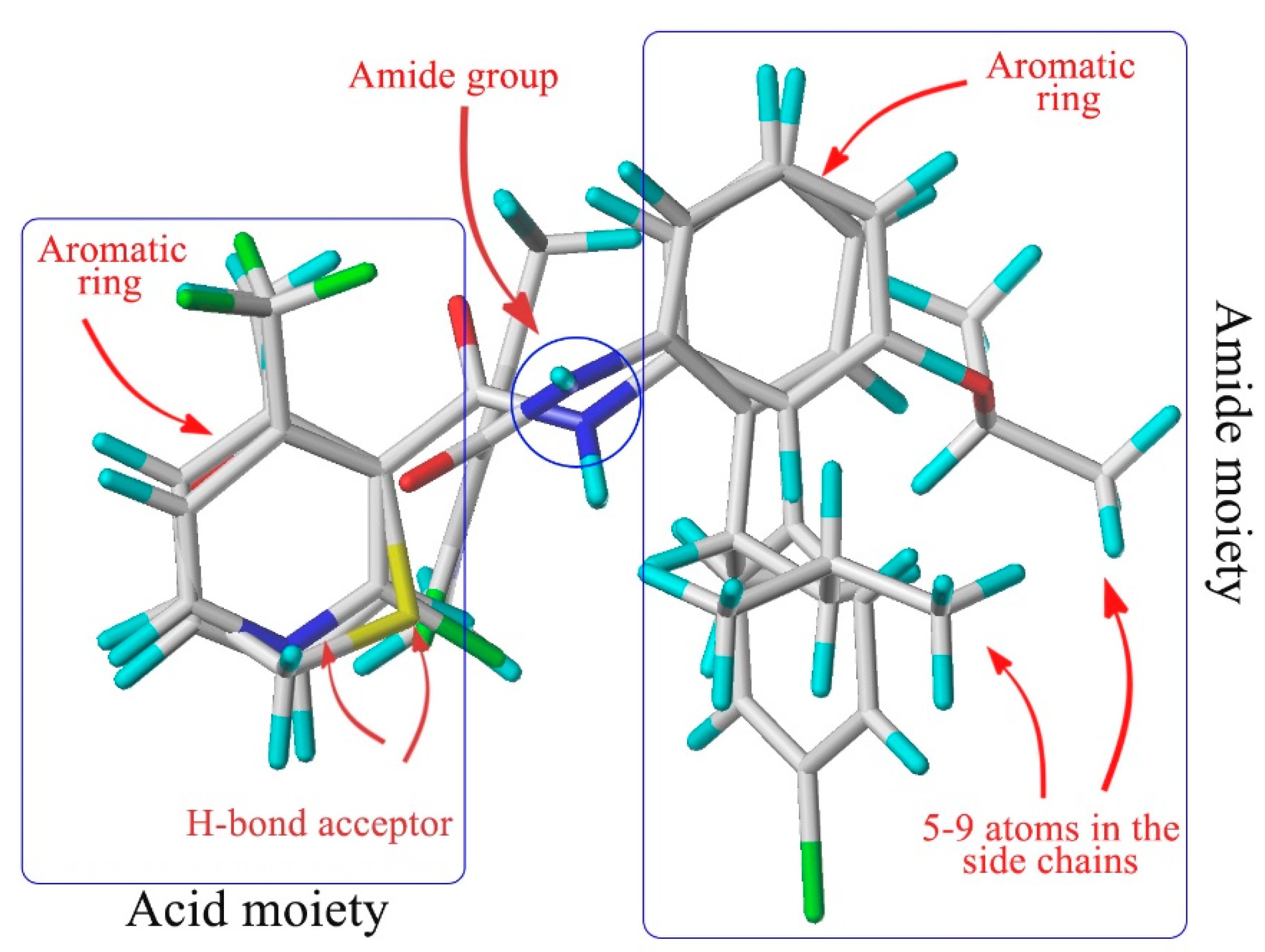
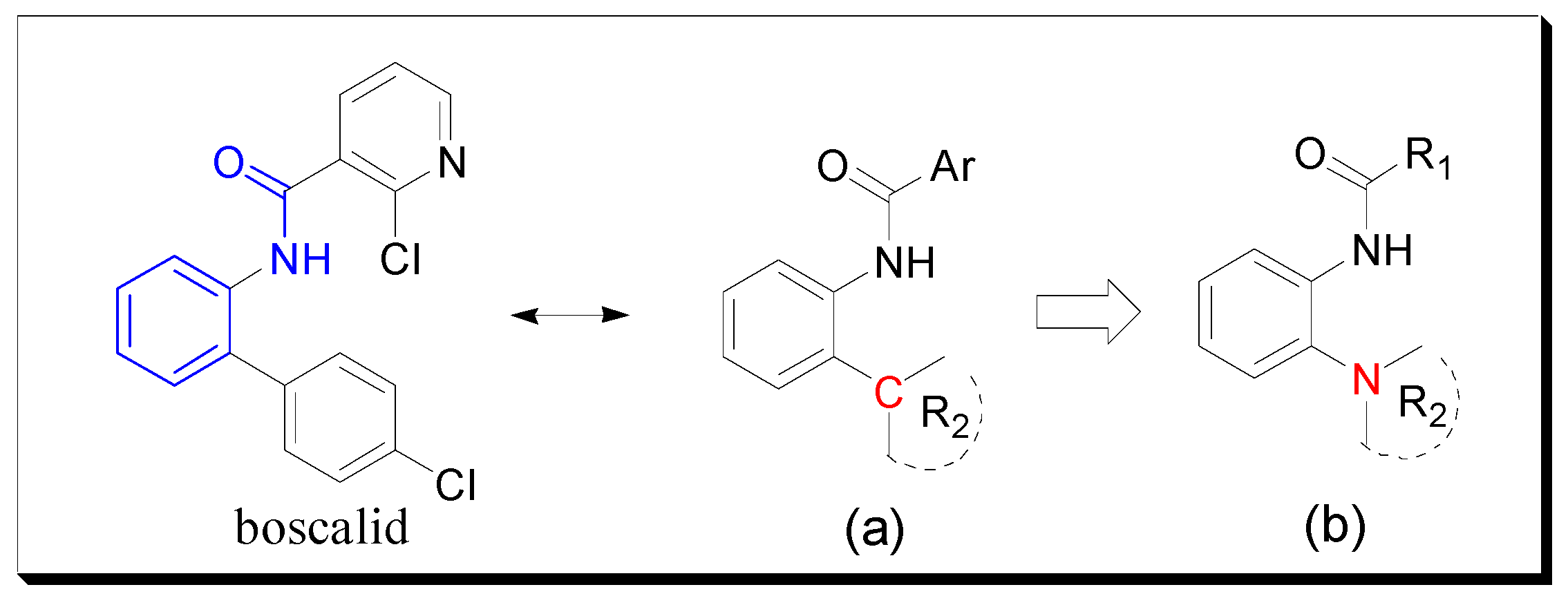
2. Results and Discussion
2.1. Synthesis of Compounds

2.2. In Vitro Antifungal Activity
| Compd. | Inhibition rate a (%) | |||||
|---|---|---|---|---|---|---|
| W. A. b | B. B. | R. S. | P. I. | P. A. | B. C. | |
| 1a | 11.11 | 30.53 | 17.56 | 33.15 | 21.17 | 12.50 |
| 1b | 39.84 | 48.09 | 59.92 | 43.67 | 37.62 | 26.22 |
| 1c | 60.43 | 27.10 | 75.19 | 18.89 | 74.14 | 79.27 |
| 1e | 24.66 | 61.83 | 55.73 | 35.83 | 51.89 | 43.29 |
| 1f | 56.10 | 55.98 | 47.71 | 42.50 | 60.58 | 31.30 |
| 1h | 44.99 | 57.25 | 67.75 | 41.33 | 48.48 | 44.51 |
| 1i | 51.22 | 54.71 | 74.24 | 45.07 | 42.89 | 51.83 |
| 2a | 8.94 | 6.11 | 43.51 | 17.72 | 32.22 | 12.23 |
| 2b | 46.61 | 25.70 | 51.53 | 41.33 | 15.27 | 14.23 |
| 2d | 19.24 | 16.79 | 21.95 | 30.11 | 24.58 | 14.23 |
| 2e | 23.31 | 11.20 | 39.69 | 27.07 | 49.41 | * |
| 2j | 15.99 | * | 50.95 | 30.81 | 46.65 | 12.50 |
| 2g | 50.95 | 40.71 | 58.78 | 34.32 | 42.27 | 21.34 |
| 2h | 42.55 | 19.59 | 55.34 | 38.99 | 43.39 | 14.02 |
| 3a | 32.93 | 31.30 | 42.56 | 31.04 | 42.89 | 11.59 |
| 3b | 48.78 | 38.93 | 49.24 | 38.29 | 39.79 | 46.34 |
| 3c | 47.97 | 56.49 | 75.57 | 36.42 | 84.17 | 49.59 |
| 3d | 35.23 | 24.94 | 39.89 | 34.32 | 34.20 | 36.38 |
| 3e | 43.63 | 28.24 | 69.08 | 33.61 | 38.18 | 23.78 |
| 3f | 57.32 | 23.66 | 43.51 | 36.65 | 49.10 | 37.20 |
| 4a | 11.38 | 18.32 | 36.45 | 17.72 | 10.61 | * |
| 4b | 37.13 | 32.57 | 50.57 | 40.63 | 44.13 | 18.90 |
| 4e | 27.10 | 32.44 | 50.95 | 30.11 | 57.79 | * |
| 4j | 50.38 | 28.04 | 33.88 | 24.05 | 18.49 | 0.00 |
| 4f | * | 37.09 | 21.04 | 31.30 | 39.73 | 63.33 |
| 4g | 13.74 | 44.75 | 31.15 | 32.32 | 39.04 | 43.75 |
| 4h | * | 47.54 | 24.59 | 29.52 | 38.58 | 20.00 |
| 5b | 41.60 | 54.97 | 43.99 | 40.20 | 40.87 | 40.94 |
| 5f | 35.11 | 52.65 | 60.11 | 32.32 | 40.87 | 49.58 |
| 5h | 37.40 | 47.42 | 55.46 | 31.30 | 42.47 | 48.75 |
| Boscalid | 71.54 | 58.58 | 78.73 | 56.50 | 81.10 | 88.60 |
| Pythium aphanidermatum | Rhizoctoniasolani | |||||||
|---|---|---|---|---|---|---|---|---|
| No. | EC50 | pEC50 | Predicted pEC50 | EC50 | pEC50 | Predicted pEC50 | ||
| (μg·mL−1) | CoMFA | CoMSIA | (μg·mL−1) | CoMFA | CoMSIA | |||
| 1a | 114.20 | 3.94 | 4.002 | 3.964 | 141.25 | 3.85 | 3.880 | 3.810 |
| 1b | 74.90 | 4.13 | 3.971 | 3.967 | 74.99 | 4.12 | 4.037 | 4.023 |
| 1c | 19.95 | 4.70 | 4.687 | 4.677 | 17.99 | 4.74 | 4.729 | 4.683 |
| 1e | 82.78 | 4.08 | 4.170 | 4.042 | 77.98 | 4.11 | 4.080 | 4.065 |
| 1f | 91.47 | 4.04 | 3.983 | 3.953 | 89.95 | 4.05 | 3.990 | 4.012 |
| 1h | 94.23 | 4.03 | 4.019 | 3.993 | 78.16 | 4.11 | 4.035 | 4.032 |
| 1i | 93.29 | 4.03 | 4.155 | 4.217 | 82.79 | 4.08 | 4.085 | 4.075 |
| 2a | 87.12 | 4.06 | 3.973 | 3.972 | 108.52 | 3.96 | 3.856 | 3.820 |
| 2b | 166.37 | 3.78 | 3.953 | 3.992 | 89.63 | 4.05 | 4.039 | 4.029 |
| 2d | 117.20 | 3.93 | 4.013 | 3.953 | 163.22 | 3.79 | 3.830 | 3.839 |
| 2e | 52.20 | 4.28 | 4.194 | 4.219 | 103.51 | 3.99 | 3.960 | 3.979 |
| 2j | 56.23 | 4.25 | 4.240 | 4.240 | 50.12 | 4.30 | 4.301 | 4.323 |
| 2g | 71.40 | 4.15 | 4.019 | 4.030 | 76.91 | 4.11 | 4.083 | 4.039 |
| 2h | 79.90 | 4.10 | 3.997 | 4.016 | 84.92 | 4.07 | 4.050 | 4.037 |
| 3a | 82.29 | 4.08 | 4.093 | 4.061 | 110.41 | 3.96 | 3.923 | 3.897 |
| 3b | 80.49 | 4.09 | 4.121 | 4.174 | 87.9 | 4.06 | 3.990 | 4.002 |
| 3c | 16.75 | 4.78 | 4.871 | 4.804 | 19.19 | 4.72 | 4.170 | 4.704 |
| 3d | 77.04 | 4.11 | 4.051 | 4.033 | 90.57 | 4.04 | 4.006 | 4.051 |
| 3e | 95.50 | 4.02 | 4.048 | 4.059 | 101.39 | 3.99 | 3.998 | 4.016 |
| 3f | 69.35 | 4.16 | 4.111 | 4.140 | 102.09 | 3.99 | 3.954 | 3.991 |
| 4a | 169.82 | 3.77 | 3.895 | 3.834 | 121.37 | 3.92 | 3.927 | 3.861 |
| 4b | 84.10 | 4.08 | 3.895 | 3.931 | 85.9 | 4.07 | 4.001 | 4.026 |
| 4e | 35.31 | 4.45 | 4.364 | 4.467 | 78.51 | 4.11 | 4.047 | 4.028 |
| 4j | 79.43 | 4.10 | 4.231 | 4.227 | 53.7 | 4.27 | 4.316 | 4.350 |
| 4f | 154.88 | 3.81 | 3.892 | 3.937 | 170.56 | 3.77 | 3.939 | 3.992 |
| 4g | 141.25 | 3.85 | 3.971 | 3.965 | 101.39 | 3.99 | 4.018 | 4.034 |
| 4h | 151.35 | 3.82 | 3.869 | 3.910 | 79.62 | 4.10 | 4.012 | 4.013 |
| 5b | 31.29 | 4.50 | 4.388 | 4.433 | 103.04 | 3.99 | 3.957 | 4.054 |
| 5f | 66.10 | 4.18 | 4.201 | 4.098 | 71.94 | 4.14 | 4.142 | 4.194 |
| 5h | 41.68 | 4.38 | 4.315 | 4.405 | 95.72 | 4.02 | 4.022 | 3.969 |
| Boscalid | 10.68 | 4.97 | 4.960 | 4.938 | 14.47 | 4.84 | 4.896 | 4.860 |
2.3. Quantitative Structure-Activity Relationship (QSAR) Analyses

2.4. Molecular Docking

3. Experimental Section
3.1. General Information
3.2. Synthesis of Compounds
3.2.1. Synthesis of Compounds I: 1-(2-Nitrophenyl) Piperidine
3.2.2. Synthesis of Compounds II: 2-(Piperidin-1-yl)aniline
3.2.3. Synthesis of Compounds III: 2-Chloro-N-(2-(piperidin-1-yl)phenyl)nicotinamide (1b)
3.3. Bioassays
3.4. QSAR Analyses
3.5. Molecular Docking
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yang, J.C.; Zhang, J.B.; Chai, B.S.; Liu, C.L. Progress of the development on the novel amides fungicides. Agrochemicals. 2008, 47, 6–9. [Google Scholar]
- Wen, F.; Zhang, H.; Yu, Z.Y.; Jin, H.; Yang, Q.; Hou, T.P. Design, synthesis and antifungal/insecticidal evaluation of novel nicotinamide derivatives. Pest. Biochem. Physiol. 2010, 98, 248–253. [Google Scholar] [CrossRef]
- Liu, C.L.; Chi, H.W.; Li, Z.N.; Ouyang, J.; Luo, Y.M. Preparation and Application of Compounds of N-(Substituted pyridyl)amide. CN Patent 1,927,838A, 14 March 2007. [Google Scholar]
- Ye, Y.H.; Ma, L.; Dai, Z.C.; Xiao, Y.; Zhang, Y.Y.; Li, D.D.; Wang, J.X.; Zhu, H.L. Synthesis and antifungal activity of nicotinamide derivatives as succinate dehydrogenase inhibitors. J. Agric. Food Chem. 2014, 62, 4063–4071. [Google Scholar] [CrossRef] [PubMed]
- Scalliet, G.; Bowler, J.; Luksch, T.; Kirchhofer-Allan, L.; Steinhauer, D.; Ward, K.; Niklaus, M.; Verras, A.; Csukai, M.; Daina, A.; et al. Mutagenesis and functional studies with succinate dehydrogenase inhibitors in the wheat pathogen mycosphaerella graminicola. PLoS One 2012, 7, e35429. [Google Scholar] [CrossRef] [PubMed]
- Ralph, S.J.; Moreno-Sanchez, R.; Neuzil, J.; Rodriguez-Enriquez, S. Inhibitors of succinate: Quinone reductase/complex ii regulate production of mitochondrial reactive oxygen species and protect normal cells from ischemic damage but induce specific cancer cell death. Pharmaceut. Res. 2011, 28, 2695–2730. [Google Scholar] [CrossRef]
- Huang, Q.C. Progress of the activity and mechanism of the amide fungicides. World Pest. 2004, 26, 23–27. [Google Scholar]
- Dehne, H.W.; Deising, D.H.; Gisi, U.; Kuck, K.H.; Russell, P.E.; Lyr, H. Modern fungicides and antifungal compounds vi. In Proceedings of the 16th International Reinhardsbrunn Symposium, Friedrichroda, Germany, 25–29 April 2010; p. 161.
- Patani, G.A.; LaVoie, E.J. Bioisosterism: A rational approach in drug design. Chem. Rev. 1996, 96, 3147–3176. [Google Scholar] [CrossRef] [PubMed]
- Lima, L.M.; Barreiro, E.J. Bioisosterism: A useful strategy for molecular modification and drug design. Curr. Med. Chem. 2005, 12, 23–49. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Beutler, A.; Snovydovych, B. Recent advances in the chemistry of indazoles. Eur. J. Org. Chem. 2008, 24, 4073–4095. [Google Scholar] [CrossRef]
- Cerecetto, H.; Gerpe, A.; González, M.; Aran, V.J.; de Ocariz, C.O. Pharmacological properties of indazole derivatives: Recent developments. Mini-Rev. Med. Chem. 2005, 5, 869–878. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.S.; Sun, G.; Cobessi, D.; Wang, A.C.; Shen, J.T.; Tung, E.Y.; Anderson, V.E.; Berry, E.A. 3-Nitropropionic acid is a suicide inhibitor of mitochondrial respiration that, upon oxidation by complex ii, forms a covalent adduct with a catalytic base arginine in the active site of the enzyme. J. Biol. Chem. 2006, 281, 5965–5972. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.J.; Zhai, Y.J.; Lou, J.Z.; Liu, M.; Pang, X.Y.; Sun, F. Thiabendazole inhibits ubiquinone reduction activity of mitochondrial respiratory complex ii via a water molecule mediated binding feature. Protein Cell 2011, 2, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Huo, X.; Zhai, Y.; Wang, A.; Xu, J.X.; Su, D.; Bartlam, M.; Rao, Z.H. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell 2005, 121, 1043–1057. [Google Scholar] [CrossRef] [PubMed]
- Avenot, H.F.; Michailides, T.J. Progress in understanding molecular mechanisms and evolution of resistance to succinate dehydrogenase inhibiting (SDHI) fungicides in phytopathogenic fungi. Crop. Prot. 2010, 29, 643–651. [Google Scholar] [CrossRef]
- Antilla, J.C.; Baskin, J.M.; Barder, T.E.; Buchwald, S.L. Copper-diamine-catalyzed N-arylation of pyrroles, pyrazoles, indazoles, imidazoles, and triazoles. J. Org. Chem. 2004, 69, 5578–5587. [Google Scholar] [CrossRef] [PubMed]
- Bavin, P.M.G. 2-Aminofluorene. Org. Synth. 1973, 5, 30. [Google Scholar]
- Yadav, V.; Gupta, S.; Kumar, R.; Singh, G.; Lagarkha, R. Polymeric peg35k-pd nanoparticles: Efficient and recyclable catalyst for reduction of nitro compounds. Synth. Commun. 2012, 42, 213–222. [Google Scholar] [CrossRef]
- Wang, M.; Gao, M.; Steele, B.L.; Glick-Wilson, B.E.; Brown-Proctor, C.; Shekhar, A.; Hutchins, G.D.; Zheng, Q.H. A new facile synthetic route to [11C]GSK189254, a selective PET radioligand for imaging of CNS histamine H3 receptor. Bioorg. Med. Chem. Lett. 2012, 22, 4713–4718. [Google Scholar] [CrossRef] [PubMed]
- Chi, H.W.; Li, Z.N.; Liu, C.L. Synthesis and bioactivity of fungicide boscalid. Fine Chem. Intermed. 2007, 37, 14–16. [Google Scholar]
- Yuan, X.Y.; Zhang, L.; Han, X.Q.; Zhou, Z.Y.; Du, S.J.; Wan, C.; Yang, D.Y.; Qin, Z.H. Synthesis and fungicidal activity of the strobilurins containing 1,3,4-thiodiazole ring. Chin. J. Org. Chem. 2014, 34, 170–177. [Google Scholar] [CrossRef]
- Duan, H.X.; Liang, D.; Yang, X.L. 3D Quantitative structure-property relationship study on cis-neonicotinoid derivatives. Acat. Chim. Sin. 2010, 68, 595–602. [Google Scholar]
- Su, W.C.; Zhou, Y.H.; Ma, Y.Q.; Wang, L.; Zhang, Z.; Rui, C.H.; Duan, H.X.; Qin, Z. N'-Nitro-2-hydrocarbylidenehydrazinecarboximidamides: Design, synthesis, crystal structure, insecticidal activity, and structure-activity relationships. J. Agric. Food Chem. 2012, 60, 5028–5034. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds are available from the authors.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, S.; Lu, H.; Yang, D.; Li, H.; Gu, X.; Wan, C.; Jia, C.; Wang, M.; Li, X.; Qin, Z. Synthesis, Antifungal Activity and QSAR of Some Novel Carboxylic Acid Amides. Molecules 2015, 20, 4071-4087. https://doi.org/10.3390/molecules20034071
Du S, Lu H, Yang D, Li H, Gu X, Wan C, Jia C, Wang M, Li X, Qin Z. Synthesis, Antifungal Activity and QSAR of Some Novel Carboxylic Acid Amides. Molecules. 2015; 20(3):4071-4087. https://doi.org/10.3390/molecules20034071
Chicago/Turabian StyleDu, Shijie, Huizhe Lu, Dongyan Yang, Hong Li, Xilin Gu, Chuan Wan, Changqing Jia, Mian Wang, Xiuyun Li, and Zhaohai Qin. 2015. "Synthesis, Antifungal Activity and QSAR of Some Novel Carboxylic Acid Amides" Molecules 20, no. 3: 4071-4087. https://doi.org/10.3390/molecules20034071





