Structural Analysis of Metabolites of Asiatic Acid and Its Analogue Madecassic Acid in Zebrafish Using LC/IT-MSn
Abstract
:1. Introduction
2. Results and Discussion
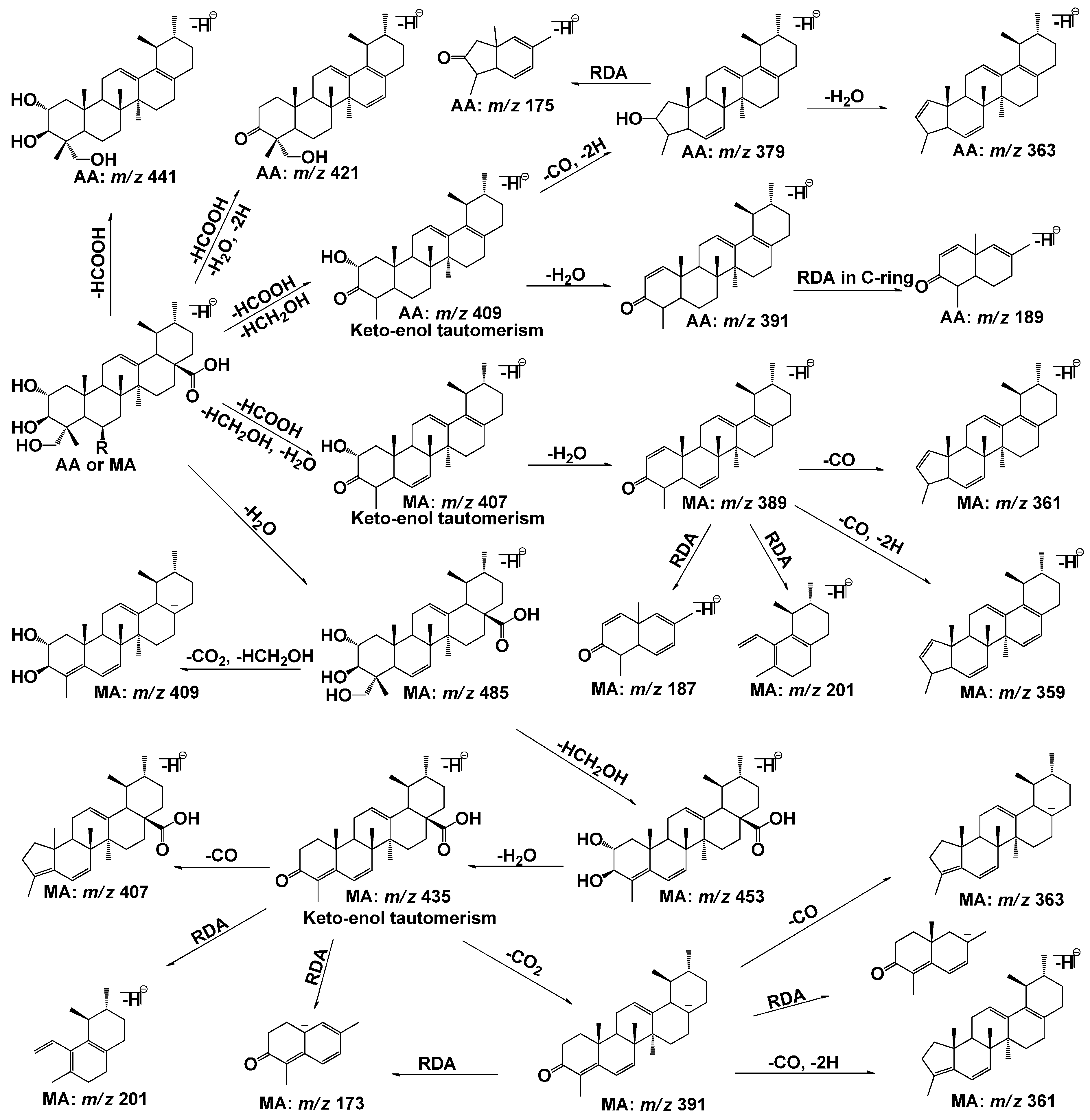
2.1. Fragmentation Pathways of AA and MA
| Metabolites | Precursor Ion ([M−H]−) | Retention Time (min) | Data-Dependent MSn Data (Collision Energy: 36%; Relative Abundance: % Base Peak) |
|---|---|---|---|
| AA | 487 | 37.57 | MS2[487]:487(35), 473(4), 443(3), 441(7), 423(4), 421(19), 409(100), 393(8), 391(10), 379(11), 153(<2) |
| MS3[487→441]:421(20), 409(100), 379(15), 233(8) | |||
| MS3[487→409]:391(50), 379(100), 375(10) | |||
| MS3[487→391]:391(100), 375(73), 373(49), 347(40), 189(48) | |||
| MS3[487→379]:379(8), 377(43), 363(100), 347(21), 225(3), 189(7), 175(13) | |||
| MS4[487→441→409]:391(28), 379(100), 226(36) | |||
| MS4[487→409→379]:377(16), 363(100), 361(60), 347(10), 225(14), 175(5) | |||
| MA | 503 | 30.51 | MS2[503]:503(96), 485(43), 465(12), 453(41), 435(33), 419(42), 407(100), 391(37), 389(97), 373(19), 371(33), 363(7), 247(2), 203(17),201(3) |
| MS3[503→485]:485(2), 465(14), 453(59), 435(100), 409(64), 391(2), 201(2) | |||
| MS3[503→407]:407(3), 405(20), 389(100), 371(12), 159(3) | |||
| MS3[503→389]:389(85), 387(18), 373(100), 371(26), 361(44), 359(79), 201(36), 187(82) | |||
| MS4[503→407→389]:389(2), 373(100) | |||
| MS4[503→485→453]:453(60), 451(35), 435(100), 417(5), 391(2), 379(4), 201(10) | |||
| MS4[503→485→435]:435(60), 407(100), 391(14), 201(4), 173(26) | |||
| MS5[503→485→453→435]:435, 417, 407, 391 | |||
| MS3[503→391]:391(100), 375(60), 363(33), 361(53), 359(3), 189(34), 187(4), 173(12) | |||
| MS4[503→391→375]:375(100), 359(38), 345(2), 189(2), 173(<2) | |||
| MS4[503→391→363]:363(100), 347(30), 223(<2), 173(30), 159(10) |
2.2. Identification of AA and Its Metabolites with Zebrafish Exposure
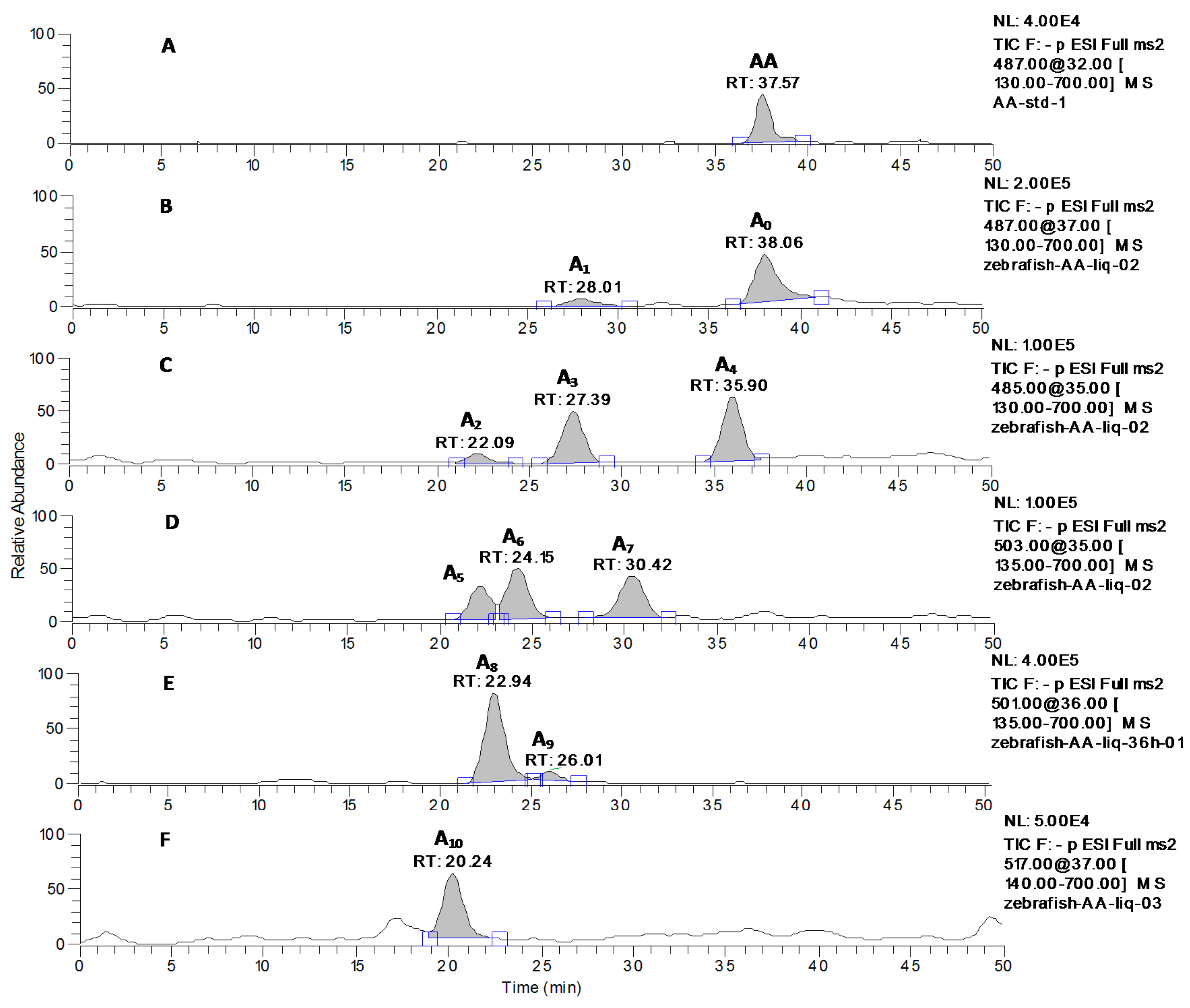
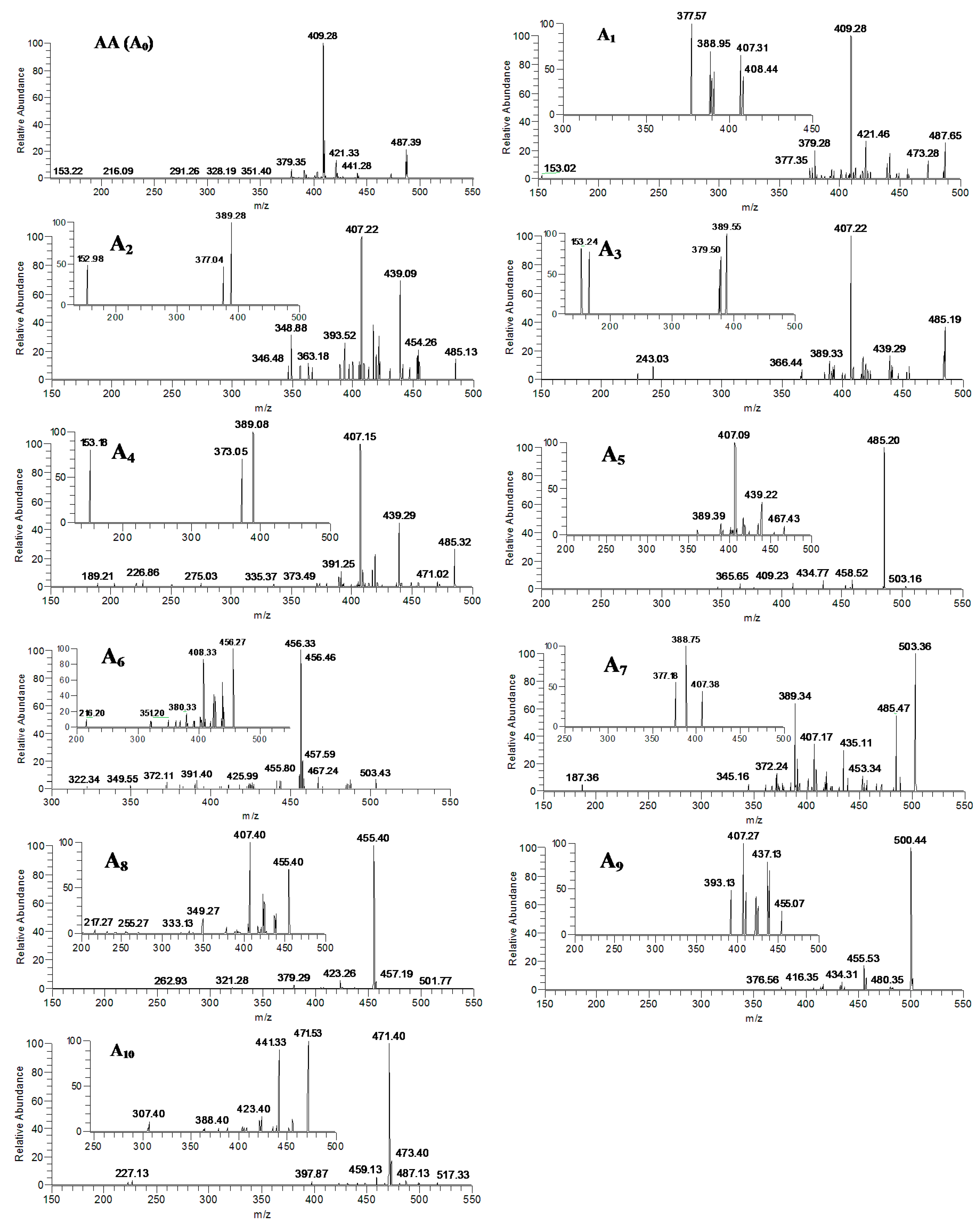
| Metabolites | Precursor Ion ([M−H]−) | Retention Time (min) | Data-Dependent MSn Data (Relative Abundance: % Base Peak) |
|---|---|---|---|
| A1 | 487 | 28.01 | MS2[487]: 487, 473, 443, 441, 421, 409(100), 379, 377, etc. |
| MS3[487→409]: 407, 391, 389, 377(100) | |||
| A2 | 485 | 22.09 | MS2[485]: 485, 455, 439, 419, 417, 407 (100), 393 etc. |
| MS3[485→455]: 455, 439, 425, 411 (100), 379 | |||
| MS3[485→439]: 439, 419, 417, 407 (100), 402, 393 | |||
| MS3[485→407]: 389(100), 377, 153 | |||
| MS4[485→407→389]: 389(100), 374, | |||
| A3 | 485 | 27.39 | MS2[485]: 485, 455, 439, 419, 417, 407 (100), 391, 389, etc. |
| MS3[485→455]: 411 (100), 391 | |||
| MS3[485→439]: 439, 407 (100), 371, 257 | |||
| MS3[485→407]: 389(100), 379, 165, 153 | |||
| MS4[485→407→389]: 389, 371(100) | |||
| A4 | 485 | 35.90 | MS2[485]: 485, 471, 455, 439, 419, 417, 407 (100), 391, 389, 373, 189 etc. |
| MS3[485→455]: 455(100), 407, 391, 389 | |||
| MS3[485→439]: 439(100), 419, 417, 407 (100) | |||
| MS3[485→407]: 389(100), 373, 153 | |||
| MS4[485→407→389]: 389(100) | |||
| A5 | 503 | 22.22 | MS2[503]: 503, 485(100), 459, 435, 409, etc. |
| MS3[503→485]: 467, 439, 419, 417, 407(100), 389 | |||
| MS4[503→485→407]: 391, 389, 377(100) | |||
| A6 | 503 | 24.15 | MS2[503]: 503, 487, 467, 457, 456(100), 443, 425, 411, 391, etc. |
| MS3[503→457]: 456(100), 438, 424, 408, 392, etc. | |||
| MS4[503→457→407]: 408(100), 407, 392, 390, 379, 377 | |||
| A7 | 503 | 30.42 | MS2[503]: 503(100), 485, 453, 435, 407, 391, 389, etc. |
| MS3[503→453]: 453, 435, 423(100) | |||
| MS3[503→407]: 407, 389(100), 377 | |||
| A8 | 501 | 22.94 | MS2[501]: 501, 457, 455(100), 423, 379, etc. |
| MS3[501→457]: 457, 455, 439, 437, 425(100), 407, 393, etc. | |||
| MS3[501→455]: 455,439, 437, 425, 423, 407(100), 379, 349 | |||
| MS3[501→423]: 423(100), 407, 405, 389, 379 | |||
| MS3[501→379]: 379(100), 361, 353, 335 | |||
| MS4[501→455→407]: 407(100), 391, 379, 377, etc. | |||
| A9 | 501 | 26.01 | MS2[501]: 501(100), 457, 455, 434, 416, 377 etc. |
| MS3[501→455]: 455,439, 437, 425, 423, 409, 407(100), 393 | |||
| A10 | 517 | 20.24 | MS2[517]: 517, 499, 487, 473, 471(100), 459, etc. |
| MS3[517→471]: 471(100), 455, 441, 423, 421, etc. | |||
| MS4[517→471→441]: 441(100), 423, 409, 393, 391, 379, 375, 363, etc. | |||
| MS4[517→471→455]: 455(100), 439, 407, 405 | |||
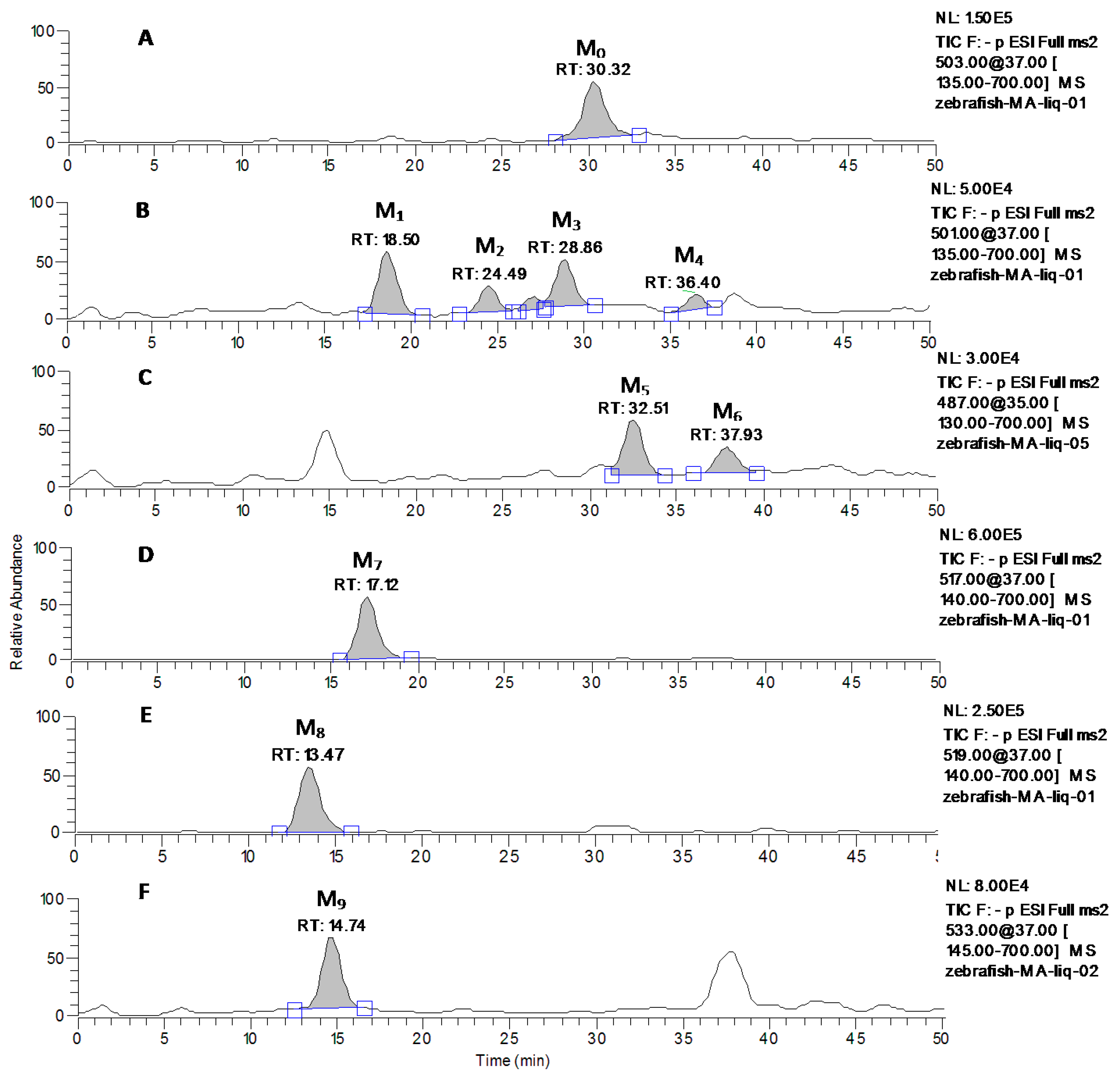
| M1 | 501 | 18.50 | MS2[501]: 501, 483(100), 457, 435, 419, 417, 407, 405, 389, 373, 361, etc. |
| MS3[501→483]: 435, 407(100), 389, 371 | |||
| MS3[501→435]: 417(100), 389 | |||
| MS3[501→407]: 389(100), 371 | |||
| MS3[501→389]: 389(100), 373, etc. | |||
| M2 | 501 | 24.49 | MS2[501]: 501, 483, 471, 455(100), 425, 407, 379, etc. |
| MS3[501→455]: 455(100), 439, 437, 425, 423, 407, 389, 379, 361, etc. | |||
| MS3[501→407]: 407, 389(100), 377 | |||
| M3 | 501 | 28.86 | MS2[501]: 501, 483(100), 457, 455, 437, 435, 419, 407,405, 389, 371, etc. |
| MS3[501→455]: 483, 453, 437, 435, 417, 407, 405, 389, 387, 363(100) | |||
| M4 | 501 | 36.40 | MS2[501]: 501, 483, 463, 437, 435, 433, 419, 407,405, 389(100), 371, etc. |
| MS3[501→389]: 389, 387(100) | |||
| M5 | 487 | 32.51 | MS2[487]: 487, 469, 457,441, 423(100), etc. |
| MS3[487→423]: 393(100), 467, 349 | |||
| M6 | 487 | 37.93 | MS2[487]: 487, 457,441, 419, 409(100), etc. |
| MS3[487→409]: 379(100), 363 | |||
| M7 | 517 | 17.12 | MS2[517]: 517, 501, 471(100), 453, 423, 407, 405, 391, etc. |
| MS3[517→471]: 453, 435, 423, 405(100), 387, 371, etc. | |||
| MS3[517→453]: 435, 423(100), 405, 387, 347, 173 | |||
| MS3[517→423]: 407, 405(100), 389, 387, 357, 341 | |||
| MS4[517→471→453]: 453, 435, 425, 423, 407, 405(100), 389,387 | |||
| MS4[517→471→423]: 423, 405(100), 387, 357 | |||
| MS4[517→471→405]: 405(100), 387, 377, 371, 357, etc. | |||
| M8 | 519 | 13.47 | MS2[519]: 519, 501(100), 483, etc. |
| MS3[519→501]: 483, 435(100), 425 | |||
| MS3[519→483]: 483, 471, 435, 417, 407, 405, 389, 371, 363, 361, etc. | |||
| MS4[519→501→483]: 465, 435(100), 407 | |||
| M9 | 533 | 14.74 | MS2[533]: 533, 489, 487(100), 469, 425, 403, etc. |
| MS3[533→489]: 489, 471, 441, 423(100), 405, 381, 365, etc. | |||
| MS3[533→487]: 487(100), 469, 457, 439, 421, 403, 363, etc. | |||
| MS4[533→489→471]:423, 405, 395, 381, 363(100) | |||
| MS4[533→487→469]: 469, 454, 439(100), 421, 403, 387, 379 |
| Description | Molecular Formula Change | m/z Change |
|---|---|---|
| Decarboxylation | −CO2 | −44 |
| Hydroxymethylene loss | −CH2O | −30 |
| Demethylation | −CH2 | −14 |
| Hydroxylation + dehydration | −2H | −2 |
| Dehydrogenation | −2H | −2 |
| Demethylation + hydroxylation | −CH2, +O | +2 |
| Methylene to ketone | −2H+O | +14 |
| Hydroxylation + dehydrogenation | −2H+O | +14 |
| Methylation | +CH2 | +14 |
| Hydroxylation | +O | +16 |
| Methyl to carboxylation | −2H+2O | +30 |
| Hydroxylation and methylation | +CH2O | +30 |
| Dihydroxylation+ dehydrogenation | +2O, −2H | +30 |
| Dihydroxylation | +2O | +32 |
| Tri-hydroxylation + dehydrogenation | +3O, −2H | +46 |
| Tri-hydroxylation | +3O | +48 |
| Glycine conjugation | +C2H3NO | +57 |
| Sulfation | +SO3 | +80 |
| Hydroxylation and sulfation | +SO4 | +96 |
| Cysteine conjugation | +C3H5NOS | +103 |
| Taurine conjugation | +C2H5NO2S | +107 |
| S,N-Acetylcysteine onjugation | +C5H7NO2S | +145 |
| Glucosidation | +C6H10O5 | +162 |
| Glucuronide conjugation | +C6H8O6 | +176 |
| Hydroxylation + glucuronide | +C6H8O7 | +192 |
| Glutathione conjugation | +C10H15N3O6S | +305 |
| Glutathione conjugation | +C10H17N3O6S | +307 |
2.3. Identification of MA and Its Metabolites with Zebrafish Exposure
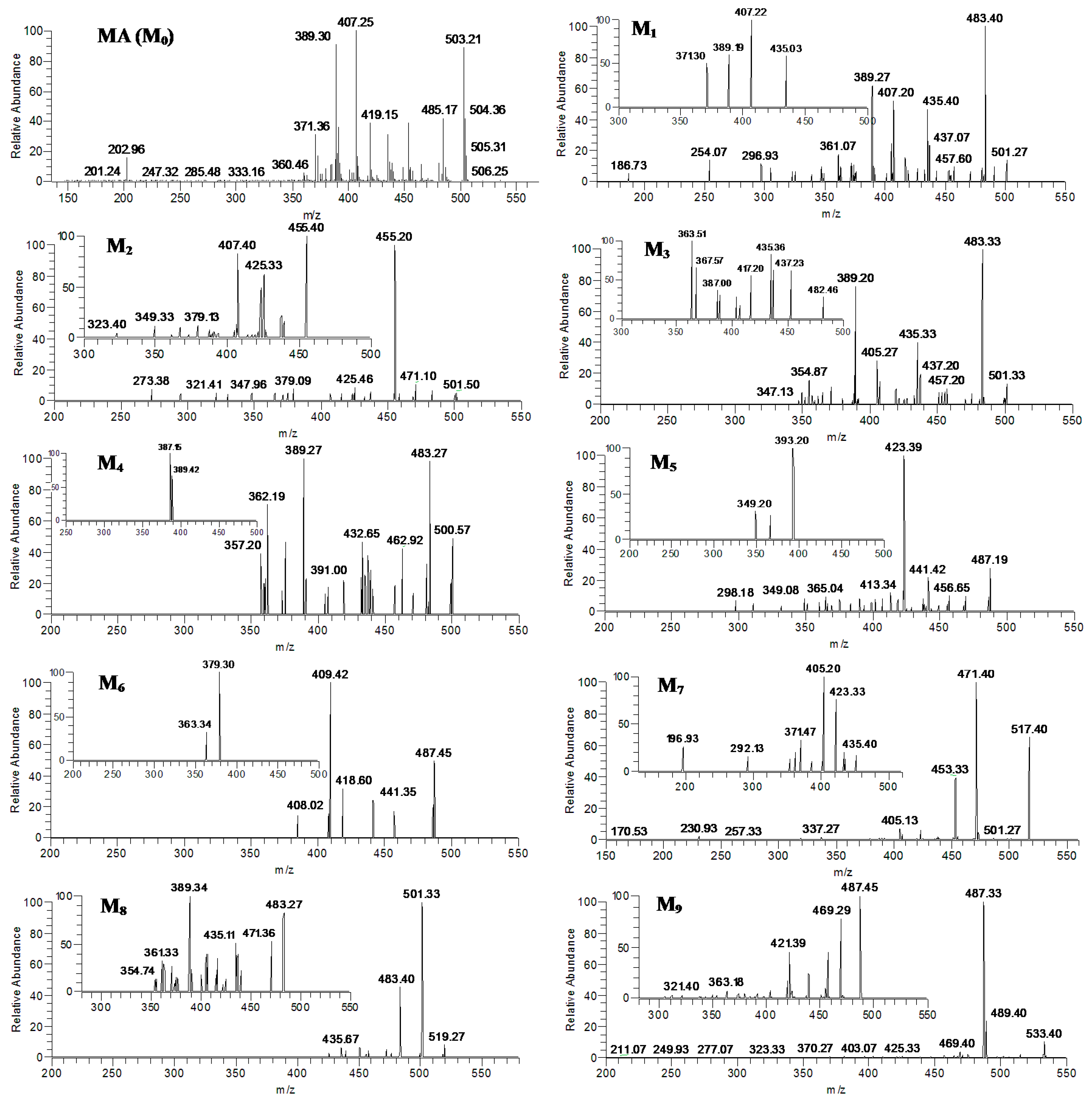
2.4. Verification of Metabolites by Authentic Standards
2.5. Discussion
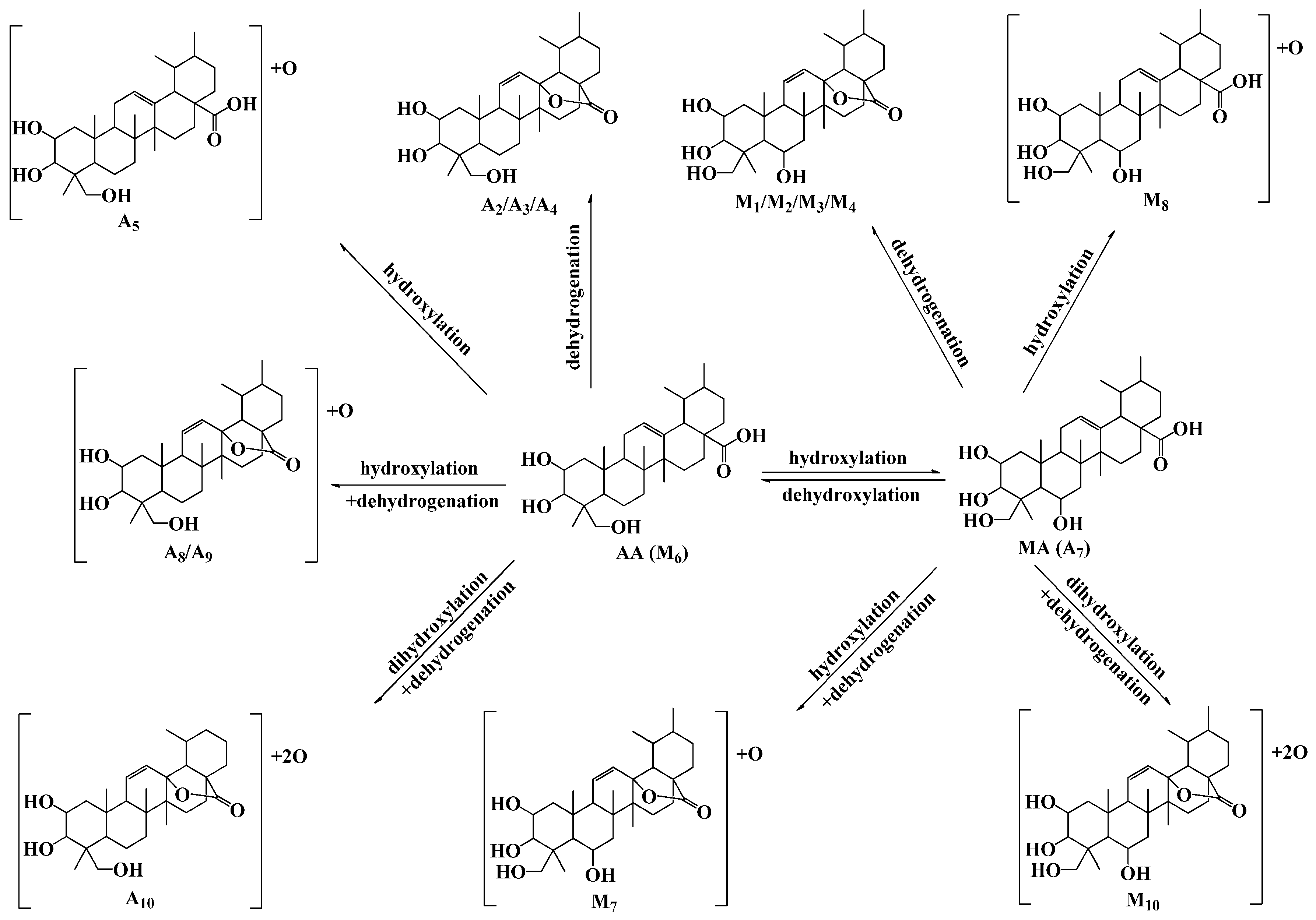
3. Experimental Section
3.1. Chemicals and Reagents
3.2. Apparatus and Analytical Conditions
3.3. Animal Protocol
3.4. Metabolism of AA and MA in Adult Zebrafish
3.5. Sample Preparation
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chinese Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China; China Medical Science and Technology Press: Beijing, China, 2010; Volume 1, p. 266. [Google Scholar]
- An, I.S.; An, S.; Choe, T.Β.; Kang, S.Μ.; Lee, J.H.; Park, I.C.; Jin, Y.W.; Lee, S.J.; Bae, S. Centella asiatica protects against UVB-induced HaCaT keratinocyte damage through microRNA expression changes. Int. J. Mol. Med. 2012, 30, 1349–1356. [Google Scholar]
- Hong, S.S.; Kim, J.H.; Li, H.; Shim, C.K. Advanced formulation and pharmacological activity of hydrogel of the titrated extract of C. asiatica. Arch. Pharm. Res. 2005, 28, 502–508. [Google Scholar] [CrossRef]
- Cho, M.K.; Sung, M.A.; Kim, D.S.; Park, H.G.; Jew, S.S.; Kim, S.G. 2-Oxo-3,23-isopropylidene-asiatate (AS2006A), a wound-healing asiatate derivative, exerts anti-inflammatory effect by apoptosis of macrophages. Int. Immunopharmacol. 2003, 3, 1429–1437. [Google Scholar] [CrossRef] [PubMed]
- Won, J.H.; Shin, J.S.; Park, H.J.; Jung, H.J.; Koh, D.J.; Jo, B.G.; Lee, J.Y.; Yun, K.; Lee, K.T. Anti-inflammatory effects of madecassic acid via the suppression of NF-B pathway in LPS-induced RAW 246.7 macrophage cells. Planta Med. 2010, 76, 251–257. [Google Scholar] [CrossRef]
- Yadav, V.R.; Prasad, S.; Sung, B.; Kannappan, R.; Aggarwal, B.B. Targeting inflammatory pathways by triterpenoids for prevention and treatment of cancer. Toxins 2010, 2, 2428–2466. [Google Scholar] [CrossRef] [PubMed]
- Yun, K.J.; Kim, J.Y.; Kim, J.B.; Lee, K.W.; Jeong, S.Y.; Park, H.J.; Jung, H.J.; Cho, Y.W.; Yun, K.; Lee, K.T. Inhibition of LPS-induced NO and PGE2 production by asiatic acid via NF-kappa B inactivation in RAW 264.7 macrophages: Possible involvement of the IKK and MAPK pathways. Int. Immunopharmacol. 2008, 8, 431–441. [Google Scholar]
- Maulidiani; Abas, F.; Khatib, A.; Shaari, K.; Lajisa, N.H. Chemical characterization and antioxidant activity of three medicinal Apiaceae species. Ind. Crop. Prod. 2014, 55, 238–247. [Google Scholar]
- Ramachandran, V.; Saravanan, R. Asiatic acid prevents lipid peroxidation and improves antioxidant status in rats with streptozotocin-induced diabetes. J. Funct. Foods 2013, 5, 1077–1087. [Google Scholar] [CrossRef]
- Xiong, Y.; Ding, H.; Xu, M.; Gao, J. Protective effects of asiatic acid on rotenone- or H2O2-induced injury in SH-SY5Y cells. Neurochem. Res. 2009, 34, 746–754. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.L.; Kuo, P.L.; Lin, L.T.; Lin, C.C. Asiatic acid, a triterpene, induces apoptosis and cell cycle arrest through activation of extracellular signal-regulated kinase and p38 mitogen-activated protein kinase pathways in human breast cancer cells. J. Pharmacol. Exp. Ther. 2005, 313, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Park, B.C.; Bosire, K.O.; Lee, E.S.; Lee, Y.S.; Kim, J.A. Asiatic acid induces apoptosis in SK-MEL-2 human melanoma cells. Cancer Lett. 2005, 218, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Salvador, J.A.R.; Moreira, V.M.; Goncalves, B.M.F.; Leal, A.S.; Jing, Y. Ursane-type pentacyclic triterpenoids as useful platforms to discover anticancer drugs. Nat. Prod. Rep. 2012, 29, 1463–1479. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, R.G.; Senut, M.C.; Zemke, D.; Min, J.; Frenkel, M.B.; Greenberg, E.J.; Yu, S.W.; Ahn, N.; Goudreau, J.; Kassab, M.; et al. Asiatic acid, a pentacyclic triterpene from Centella asiatica, is neuroprotective in a mouse model of focal cerebral ischemia. J. Neurosci. Res. 2009, 87, 2541–2550. [Google Scholar]
- Xiong, Y.Y.; Ding, H.Q.; Xu, M.F.; Gao, J. Neuroprotective effects of Asiatic Acid against Parkinson-like injury of SH-SY5Y cells and motor deficient of mutant drosophilas. J. Neurochem. 2010, 115, 55–55. [Google Scholar]
- Zhang, X.N.; Wu, J.Y.; Dou, Y.; Xia, B.B.; Rong, W.B.; Rimbach, G.; Lou, Y.J. Asiatic acid protects primary neurons against C2-ceramide-induced apoptosis. Eur. J. Pharmacol. 2012, 679, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Siu, S.O.; Li, S.; Chu, I.K.; Kwan, Y.W.; Chan, S.W.; Leung, G.P.; Yan, R.; Lee, S.M. Metabolism of calycosin, an isoflavone from Astragali Radix, in zebrafish larvae. Xenobiotica 2012, 42, 294–303. [Google Scholar] [CrossRef] [PubMed]
- Crawford, A.D.; Esguerra, C.V.; de Witte, P.A.M. Fishing for drugs from nature: Zebrafish as a technology platform for natural product discovery. Planta Med. 2008, 74, 624–632. [Google Scholar] [CrossRef] [PubMed]
- Scornaienchi, M.L.; Thornton, C.; Willett, K.L.; Wilson, J.Y. Functional differences in the cytochrome P450 1 family enzymes from zebrafish (Danio rerio) using heterologously expressed proteins. Arch. Biochem. Biophys. 2010, 502, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Thompson, E.D.; Burwinkel, K.E.; Chava, A.K.; Notch, E.G.; Mayer, G.D. Activity of Phase I and Phase II enzymes of the benzo[a]pyrene transformation pathway in zebrafish (Danio rerio) following waterborne exposure to arsenite. Comp. Biochem. Phys. C 2010, 152, 371–378. [Google Scholar]
- Trickler, W.J.; Guo, X.; Cuevas, E.; Ali, S.F.; Paule, M.G.; Kanungo, J. Ketamine attenuates cytochrome p450 aromatase gene expression and estradiol-17β levels in zebrafish early life stages. J. Appl. Toxicol. 2014, 34, 480–488. [Google Scholar] [CrossRef] [PubMed]
- Almeida, D.V.; Nornberg, B.F.; Geracitano, L.A.; Barros, D.M.; Monserrat, J.M.; Marins, L.F. Induction of phase II enzymes and hsp70 genes by copper sulfate through the electrophile-responsive element (EpRE): Insights obtained from a transgenic zebrafish model carrying an orthologous EpRE sequence of mammalian origin. Fish Physiol. Biochem. 2010, 36, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.A.; Bhuiyan, S.; Liu, M.Y.; Sugahara, T.; Sakakibara, Y.; Suiko, M.; Yasuda, S.; Kakuta, Y.; Kimura, M.; Williams, F.E.; et al. Zebrafish as a model for the study of the phase II cytosolic sulfotransferases. Curr. Drug Metab. 2010, 11, 538–546. [Google Scholar]
- Christen, V.; Fent, K. Tissue-, sex- and development-specific transcription profiles of eight UDP-glucuronosyltransferase genes in zebrafish (Danio rerio) and their regulation by activator of aryl hydrocarbon receptor. Aquat. Toxicol. 2014, 150, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.F.; Qi, L.W.; Zhou, J.L.; Li, P. Structural characterization and identification of oleanane-type triterpene saponins in Glycyrrhiza uralensis Fischer by rapid-resolution liquid chromatography coupled with time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2010, 24, 3261–3270. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Qin, J.J.; Su, J.; Lin, S.; Huang, Y.; Jin, H.Z.; Zhang, W.D. Identification and structural characterization of dimeric sesquiterpene lactones in Inula japonica Thunb by high-performance liquid chromatography/electrospray ionization with multi-stage mass spectrometry. Rapid Commun. Mass Spectrom. 2013, 27, 2159–2169. [Google Scholar] [CrossRef] [PubMed]
- Hamish, M.; Ester, S.B.F.; Alison, N.H.; Anita, Q. Negative ion ESI-MS analysis of natural yellow dye flavonoids-An isotopic labelling study. Int. J. Mass Spectrom. 2009, 284, 57–65. [Google Scholar] [CrossRef]
- Lee, Y.T.; Fong, T.H.; Chen, H.M.; Chang, C.Y.; Wang, Y.H.; Chern, C.Y.; Chen, Y.H. Toxicity assessments of chalcone and some synthetic chalcone analogues in a zebrafish model. Molecules 2014, 19, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.L.; Wang, G.Q.; Yang, L.M.; Huang, Z.B.; Zhang, W.Q.; Yu, L.Z. Endotoxin molecule lipopolysaccharide-induced zebrafish inflammation model: A novel screening method for anti-inflammatory drugs. Molecules 2014, 19, 2390–2409. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.X.; Hao, H.P.; Wang, G.J.; Tu, P.F.; Jiang, Y.; Liang, Y.; Dai, L.; Yang, H.; Lai, L.; Zheng, C.N.; et al. An approach to identifying sequential metabolites of a typical phenylethanoid glycoside, echinacoside, based on liquid chromatography-ion trap-time of flight mass spectrometry analysis. Talanta 2009, 572–580. [Google Scholar]
- Clarke, N.J.; Rindgen, D.; Korfmacher, W.A.; Cox, K.A. Systematic LC/MS metabolite identification in drug discovery. Anal. Chem. 2001, 73, 430A–439A. [Google Scholar] [CrossRef] [PubMed]
- Tozuka, Z.; Kaneko, H.; Shiraga, T.; Mitani, Y.; Beppu, M.; Terashita, S.; Kawamura, A.; Kagayama, A. Strategy for structural elucidation of drugs and drug metabolites using (MS)n fragmentation in an electrospray ion trap. J. Mass Spectrom. 2003, 38, 793–808. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.L.; Jing, W.H.; Chen, Y.G.; Yuan, Y.F.; Yan, R.; Wang, Y.T. H-1 nuclear magnetic resonance based-metabolomic characterization of Peucedani Radix and simultaneous determination of praeruptorin A and praeruptorin B. J. Pharmaceut. Biomed. 2014, 93, 86–94. [Google Scholar] [CrossRef]
- Feng, X.; Zou, Z.M.; Fu, S.B.; Sun, L.Z.; Su, Z.H.; Sun, D.A. Microbial oxidation and glucosidation of echinocystic acid by Nocardia coralline. J. Mol. Catal. B: Enzym. 2010, 66, 219–223. [Google Scholar] [CrossRef]
- Feng, X.; Luan, J.; Guo, F.F.; Li, D.P.; Chu, Z.Y. Microbial transformation of maslinic acid by Cunninghamella blakesleana. J. Mol. Catal. B: Enzym. 2012, 82, 127–130. [Google Scholar] [CrossRef]
- Martinez, A.; Rivas, F.; Perojil, A.; Parra, A.; Garcia-Granados, A; Fernandez-Vivas, A. Biotransformation of oleanolic and maslinic acids by Rhizomucor miehei. Phytochemistry 2013, 94, 229–237. [Google Scholar]
- Fu, S.B.; Yang, J.S.; Cui, J.L.; Sun, D.A. Biotransformation of ursolic acid by Syncephalastrum racemosum CGMCC 3.2500 and anti-HCV activity. Fitoterapia 2013, 86, 123–128. [Google Scholar]
- He, W.N.; Dai, J.G.; Ye, M.; Wu, L.J.; Guo, D.A. Microbial transformation of asiatic acid by Alternaria longipes. J. Asian Nat. Prod. Res. 2010, 12, 760–764. [Google Scholar]
- Huang, F.X.; Yang, W.Z.; Ye, F.; Tian, J.Y.; Hu, H.B.; Feng, L.M.; Guo, D.A.; Ye, M. Microbial transformation of ursolic acid by Syncephalastrum racemosum (Cohn) Schroter AS 3.264. Phytochemistry 2012, 82, 56–60. [Google Scholar] [CrossRef]
- Zhang, D.L.; Zhu, M.S.; Humphreys, W.G. Drug Metabolism in Drug Design and Development: Basic Concepts and Practice; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2007; pp. 319–359. [Google Scholar]
- Wu, G.W.; Gao, J.M.; Shi, X.W.; Zhang, Q.; Wei, S.P.; Ding, K. Microbial transformations of diosgenin with the white-rot basidiomycete Coriolus versicolor. J. Nat. Prod. 2011, 74, 2095–2101. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.J.; Li, P.; Fan, H.W.; Peng, Y.R.; Liu, W.; Wang, C.M.; Shu, L.; Jia, X.B. Metabolism of Tanshinone IIA, Cryptotanshinone and Tanshinone I from Radix Salvia Miltiorrhiza in Zebrafish. Molecules 2011, 16, 6621–6633. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds are available from the authors.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, B.; Bai, L.; Li, X.; Xiong, J.; Xu, P.; Xue, M. Structural Analysis of Metabolites of Asiatic Acid and Its Analogue Madecassic Acid in Zebrafish Using LC/IT-MSn. Molecules 2015, 20, 3001-3019. https://doi.org/10.3390/molecules20023001
Xia B, Bai L, Li X, Xiong J, Xu P, Xue M. Structural Analysis of Metabolites of Asiatic Acid and Its Analogue Madecassic Acid in Zebrafish Using LC/IT-MSn. Molecules. 2015; 20(2):3001-3019. https://doi.org/10.3390/molecules20023001
Chicago/Turabian StyleXia, Binbin, Lu Bai, Xiaorong Li, Jie Xiong, Pinxiang Xu, and Ming Xue. 2015. "Structural Analysis of Metabolites of Asiatic Acid and Its Analogue Madecassic Acid in Zebrafish Using LC/IT-MSn" Molecules 20, no. 2: 3001-3019. https://doi.org/10.3390/molecules20023001





