Drug Discovery of Host CLK1 Inhibitors for Influenza Treatment
Abstract
:1. Introduction
2. Results
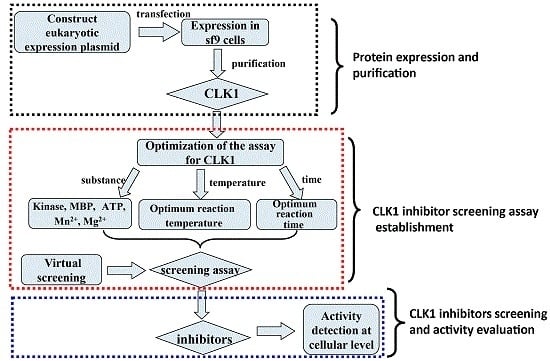
2.1. Construction of Recombined CLK1 Baculovirus, Protein Expression, Identification and Purification


2.2. Establishment of Drug Screening Assay for CLK1 Inhibitors


2.3. Rational Screening of CLK1 Inhibitors



2.4. In Vitro Anti-Influenza Viral Activities of CLK1 Inhibitors
| Samples | Name | CC50 a | CC0 b | Pre-Treatment Assay | Simultaneous Treatment Assay | Post-Treatment Assay | |||
|---|---|---|---|---|---|---|---|---|---|
| μg/mL | EC50 (μg/mL)c | SI d | EC50 (μg/mL) | SI | EC50 (μg/mL) | SI | |||
| J10688 | Clypearin | >200 | 100 | 0.75 ± 0.13 | >266.67 | 1.2 ± 0.28 | >166.67 | 18.08 ± 3.02 | >11.06 |
| J12098 | Corilagin | 153.54 | 30 | 3.99 ± 3.72 | 38.48 | 2.00 ± 2.22 | 76.77 | 5.91 ± 1.44 | 25.98 |
| J14848 | Pinosylvin | 18.26 | 10 | 7.54 ± 5.48 | 2.42 | 5.28 ± 2.45 | 3.46 | 6.65 ± 4.3 | 3.23 |
| J12133 | Chebulanin | 90.27 | 30 | 15.29 ± 10.35 | 5.90 | 9.84 ± 0.44 | 9.17 | 21.11 ± 11.15 | 4.28 |
| J25986 | Propyl gallate | 182.00 | 30 | 15.94 ± 6.94 | 11.42 | 12.88 ± 8.06 | 14.13 | 3.42 ± 0.2 | 53.22 |
| J20353 | Hispidulin | 60.35 | 30 | 18.89 ± 5.1 | 3.19 | 5.26 ± 2.48 | 11.47 | >30 | ND |
| J21548 | Norwogonin | 91.79 | 30 | >30 | ND e | >30 | ND | >30 | ND |
| J11534 | - | 175.31 | 30 | >30 | ND e | >30 | ND | >30 | ND |
| J10610 | Kaempferol | 68.50 | 30 | 5.16 ± 2.4 | 13.28 | 12.34 ± 5.89 | 5.55 | 23.3 ± 1.08 | 2.94 |
| J20725 | Luteolin | 24.86 | 10 | >10 | ND | >10 | ND | 5.74 ± 0.54 | 4.33 |
| J10857 | - | 77.72 | 30 | 18.43 ± 12.99 | 4.22 | 16.27 ± 2.29 | 4.78 | 14.67 ± 5.25 | 5.30 |
| J14528 | Isorhapontigenin | 83.37 | 30 | 5.98 ± 2.3 | 13.94 | >30 | ND | >30 | ND |
| J14080 | Epigallocatechin gallate | 71.10 | 30 | 10.19 ± 6.22 | 6.98 | 14.86 ± 1.81 | 4.78 | >30 | ND |
| Methyl gallate | Methyl gallate | 69.50 | 30 | >30 | ND e | >30 | ND | >30 | ND |
| Genistein | Genistein | 75.60 | 30 | >30 | ND | >30 | ND | >30 | ND |
| Ribavirin | Ribavirin | >200 | 100 | 4.36 ± 0.99 | >45.87 | 10.32 ± 2.38 | >19.38 | 13.78 ± 3.47 | >14.51 |
| TG003 | TG003 | 121.50 | 30 | 5.09 ± 0.78 | 23.87 | 15.75 ± 3.49 | 7.71 | 3.56 ± 1.47 | 34.13 |
3. Experimental Section
3.1. Cell Culture
3.2. Virus
3.3. Compounds
3.4. Construction of the Recombinant Human CLK1 Baculovirus
3.5. Preparation and Purification of Recombinant Human His-CLK1
3.6. Optimization of the Assay for CLK1
3.7. Rational Screening of CLK1 Inhibitors
3.8. Cytotoxicity Assay
3.9. Cytopathic Effect (CPE) Reduction Assay
3.10. Statistical Analysis
4. Discussion
| Kinase Reaction System | Authors’ Experiment | Menegay’s Experiment |
|---|---|---|
| recombinant CLK1 | 2.98 ng in 20 μL | 0.5 μg in 50μL |
| myelin basic protein(MBP) | 1 μg in 20 μL | 1 μg in 50 μL |
| MgCl2 | 10 mM | 10 mM |
| MnCl2 | 1 mM | 2 mM |
| ATP | 1 μM | 10 μM ([γ-32P]) |
| reaction temperature | room temperature | room temperature |
| reaction time | 90 min | 20 min |
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interests
References
- Lamb, R.A.; Choppin, P.W. The gene structure and replication of influenza virus. Ann. Rev. Biochem. 1983, 52, 467–506. [Google Scholar] [PubMed]
- Osterhaus, A.; Brooks, A.; Broberg, E.; Macintyre, R.; Capua, I. Why should influenza be a public health priority? Vaccine 2015. [Google Scholar] [CrossRef] [PubMed]
- Dong, G.; Peng, C.; Luo, J.; Wang, C.; Han, L.; Wu, B.; Ji, G.; He, H. Adamantane-resistant influenza a viruses in the world (1902–2013): Frequency and distribution of M2 gene mutations. PLoS ONE 2015, 10, e0119115. [Google Scholar] [CrossRef] [PubMed]
- Li, T.C.; Chan, M.C.; Lee, N. Clinical Implications of Antiviral Resistance in Influenza. Viruses 2015, 7, 4929–4944. [Google Scholar] [CrossRef] [PubMed]
- Aubol, B.E.; Plocinik, R.M.; Keshwani, M.M.; McGlone, M.L.; Hagopian, J.C.; Ghosh, G.; Fu, X.D.; Adams, J.A. N-terminus of the protein kinase CLK1 induces SR protein hyperphosphorylation. Biochem. J. 2014, 462, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Hao, L.; Sakurai, A.; Watanabe, T.; Sorensen, E.; Nidom, C.A.; Newton, M.A.; Ahlquist, P.; Kawaoka, Y. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 2008, 454, 890–893. [Google Scholar] [CrossRef] [PubMed]
- Konig, R.; Stertz, S.; Zhou, Y.; Inoue, A.; Hoffmann, H.H.; Bhattacharyya, S.; Alamares, J.G.; Tscherne, D.M.; Ortigoza, M.B.; Liang, Y.; et al. Human host factors required for influenza virus replication. Nature 2010, 463, 813–817. [Google Scholar] [CrossRef] [PubMed]
- Karlas, A.; Machuy, N.; Shin, Y.; Pleissner, K.P.; Artarini, A.; Heuer, D.; Becker, D.; Khalil, H.; Ogilvie, L.A.; Hess, S.; et al. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 2010, 463, 818–822. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Nakajima, T.; Shibata, H.; Arimura, T.; Yasunami, M.; Kimura, A. A novel link of HLA locus to the regulation of immunity and infection: NFKBIL1 regulates alternative splicing of human immune-related genes and influenza virus M gene. J. Autoimmune 2013, 47, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Muraki, M.; Ohkawara, B.; Hosoya, T.; Onogi, H.; Koizumi, J.; Koizumi, T.; Sumi, K.; Yomoda, J.; Murray, M.V.; Kimura, H.; et al. Manipulation of alternative splicing by a newly developed inhibitor of Clks. J. Biol. Chem. 2004, 279, 24246–24254. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Carter, G.; Romero, J.; Gower, K.M.; Watson, J.; Patel, N.A.; Cooper, D.R. Clk/STY (cdc2-like kinase 1) and Akt regulate alternative splicing and adipogenesis in 3T3-L1 pre-adipocytes. PLoS ONE 2013, 8, e53268. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, A.S.; Tanega, C.; Shen, M.; Mott, B.T.; Bougie, J.M.; Nguyen, D.T.; Misteli, T.; Auld, D.S.; Maloney, D.J.; Thomas, C.J. Potent and selective small molecule inhibitors of specific isoforms of Cdc2-like kinases (Clk) and dual specificity tyrosine-phosphorylation-regulated kinases (Dyrk). Bioorg. Med. Chem. Lett. 2011, 21, 3152–3158. [Google Scholar] [CrossRef] [PubMed]
- Fedorov, O.; Huber, K.; Eisenreich, A.; Filippakopoulos, P.; King, O.; Bullock, A.N.; Szklarczyk, D.; Jensen, L.J.; Fabbro, D.; Trappe, J.; et al. Specific CLK inhibitors from a novel chemotype for regulation of alternative splicing. Chem. Biol. 2011, 18, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Grabher, P.; Durieu, E.; Kouloura, E.; Halabalaki, M.; Skaltsounis, L.A.; Meijer, L.; Hamburger, M.; Potterat, O. Library-based discovery of DYRK1A/CLK1 inhibitors from natural product extracts. Planta Med. 2012, 78, 951–956. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.H.; Chung, T.D.; Oldenburg, K.R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen. 1999, 4, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Roy, P.; Noad, R. Use of bacterial artificial chromosomes in baculovirus research and recombinant protein expression: current trends and future perspectives. ISRN Microbiol. 2012, 2012, 628797. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.E.; Summers, M.D.; Fraser, M.J. Production of human beta interferon in insect cells infected with a baculovirus expression vector. 1983. Biotechnology 1992, 24, 434–443. [Google Scholar] [PubMed]
- Menegay, H.J.; Myers, M.P.; Moeslein, F.M.; Landreth, G.E. Biochemical characterization and localization of the dual specificity kinase CLK1. J. Cell Sci. 2000, 113, 3241–3253. [Google Scholar] [PubMed]
- Li, Y.; Leung, K.T.; Yao, F.; Ooi, L.S.; Ooi, V.E. Antiviral flavans from the leaves of Pithecellobium clypearia. J. Nat. Prod. 2006, 69, 833–835. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.J.; Luo, T.; Wu, F.; Liu, H.; Li, H.R.; Mei, Y.W.; Zhang, S.L.; Tao, J.Y.; Dong, J.H.; Fang, Y.; et al. Corilagin protects against HSV1 encephalitis through inhibiting the TLR2 signaling pathways in vivo and in vitro. Mol. Neurobiol. 2015, 52, 1547–1560. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.T.; Lin, C.Y.; Ho, D.H.; Peng, J.; Chen, Y.; Tsang, S.W.; Wong, M.; Zhang, X.J.; Zhang, M.; Bian, Z.X. Inhibitory effect of the gallotannin corilagin on dextran sulfate sodium-induced murine ulcerative colitis. J. Nat. Prod. 2013, 76, 2120–2125. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Jin, H.; Zhou, J.; Chen, L.; Lu, Y.; Ming, Y.; Yu, Y. A potential anti-tumor herbal medicine, Corilagin, inhibits ovarian cancer cell growth through blocking the TGF-beta signaling pathways. BMC Complement. Altern. Med. 2013, 13, 33. [Google Scholar] [CrossRef] [PubMed]
- Laavola, M.; Nieminen, R.; Leppanen, T.; Eckerman, C.; Holmbom, B.; Moilanen, E. Pinosylvin and monomethylpinosylvin, constituents of an extract from the knot of Pinus sylvestris, reduce inflammatory gene expression and inflammatory responses in vivo. J. Agric. Food Chem. 2015, 63, 3445–3453. [Google Scholar] [CrossRef] [PubMed]
- Koskela, A.; Reinisalo, M.; Hyttinen, J.M.; Kaarniranta, K.; Karjalainen, R.O. Pinosylvin-mediated protection against oxidative stress in human retinal pigment epithelial cells. Mol. Vis. 2014, 20, 760–769. [Google Scholar] [PubMed]
- Yatkin, E.; Polari, L.; Laajala, T.D.; Smeds, A.; Eckerman, C.; Holmbom, B.; Saarinen, N.M.; Aittokallio, T.; Makela, S.I. Novel Lignan and stilbenoid mixture shows anticarcinogenic efficacy in preclinical PC-3M-luc2 prostate cancer model. PLoS ONE 2014, 9, e93764. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Not Available.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zu, M.; Li, C.; Fang, J.-S.; Lian, W.-W.; Liu, A.-L.; Zheng, L.-S.; Du, G.-H. Drug Discovery of Host CLK1 Inhibitors for Influenza Treatment. Molecules 2015, 20, 19735-19747. https://doi.org/10.3390/molecules201119653
Zu M, Li C, Fang J-S, Lian W-W, Liu A-L, Zheng L-S, Du G-H. Drug Discovery of Host CLK1 Inhibitors for Influenza Treatment. Molecules. 2015; 20(11):19735-19747. https://doi.org/10.3390/molecules201119653
Chicago/Turabian StyleZu, Mian, Chao Li, Jian-Song Fang, Wen-Wen Lian, Ai-Lin Liu, Li-Shu Zheng, and Guan-Hua Du. 2015. "Drug Discovery of Host CLK1 Inhibitors for Influenza Treatment" Molecules 20, no. 11: 19735-19747. https://doi.org/10.3390/molecules201119653