Purification and Partial Characterization of β-Glucosidase in Chayote (Sechium edule)
Abstract
:1. Introduction
2. Results and Discussion
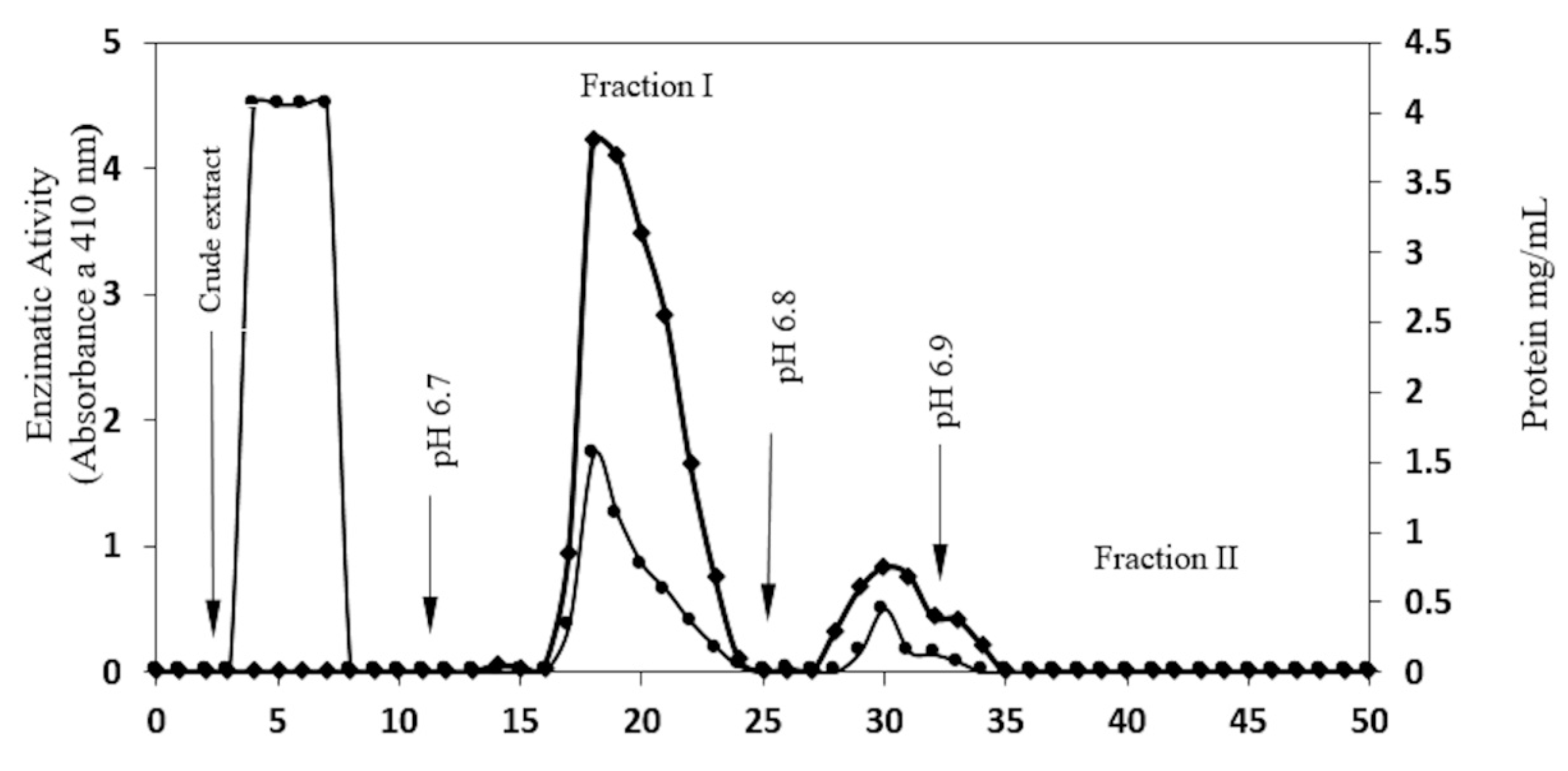
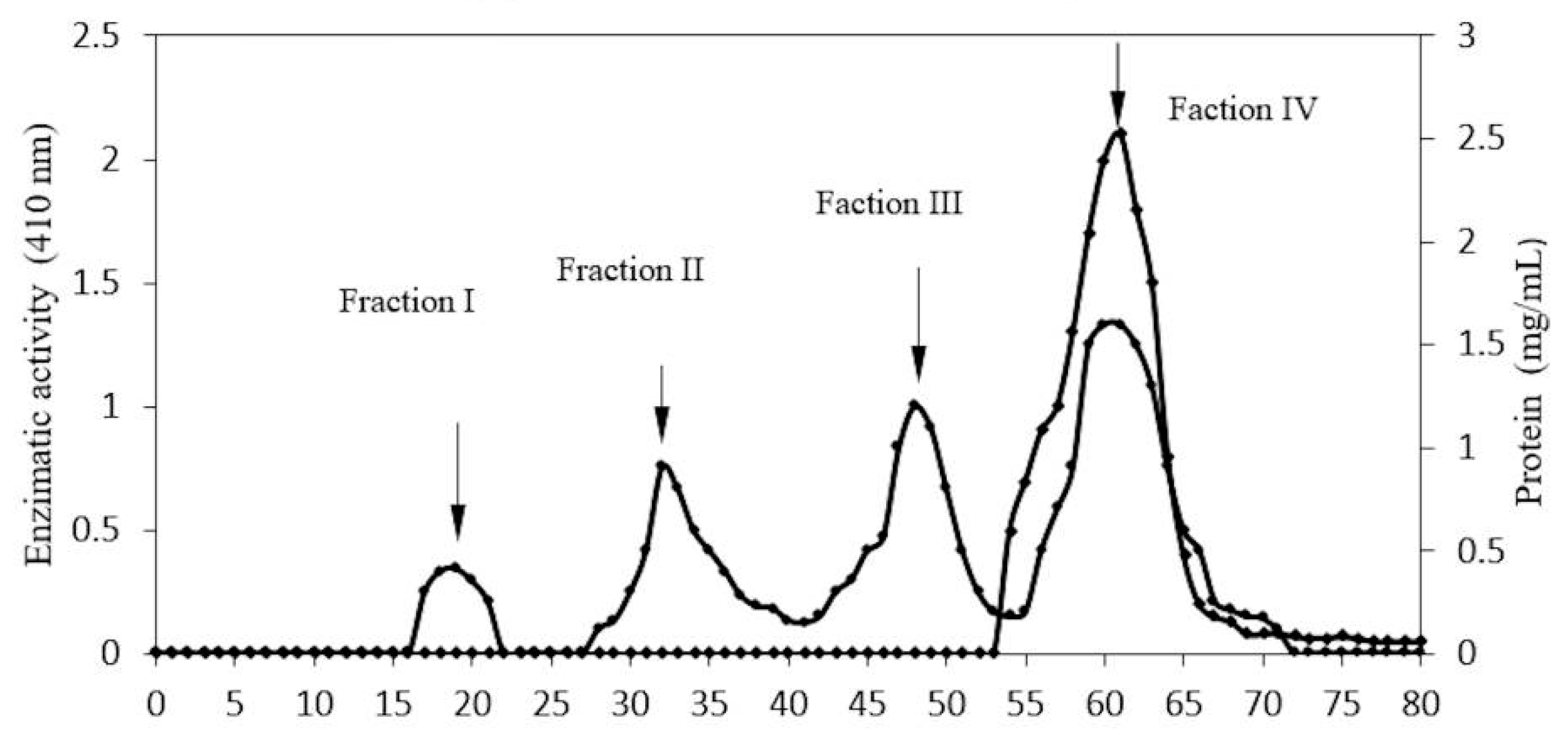
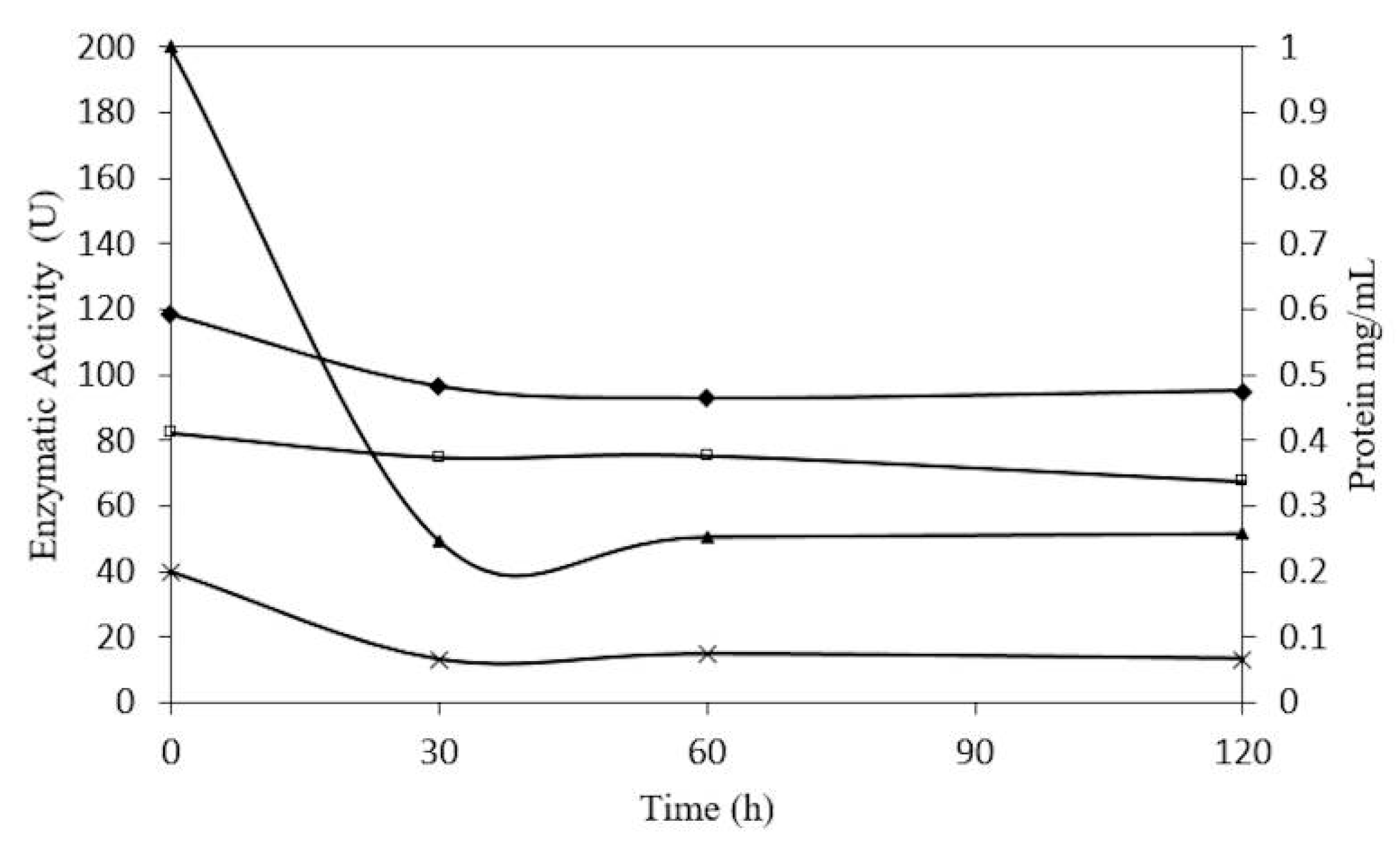
2.1. Enzyme Purification
| Fraction | Total Protein (mg) | Activity of β-Glucosidase (U) | Specific Activity (U/mg) | Purification | Yield (%) |
|---|---|---|---|---|---|
| Crude extract | 134.37 | 393.45 | 2.92 | 1 | 100 |
| Protein precipitated | 52.14 | 285.62 | 5.47 | 1.87 | 72.59 |
| CMCEL Fraction I | 12.50 | 198.81 | 15.9 | 5.44 | 50.5 |
| CMCEL fraction II | 3.16 | 128 | 40.5 | 13.86 | 32.53 |
| Sephacryl- Fraction IV | 2.46 | 155.27 | 63.11 | 21.61 | 39.46 |
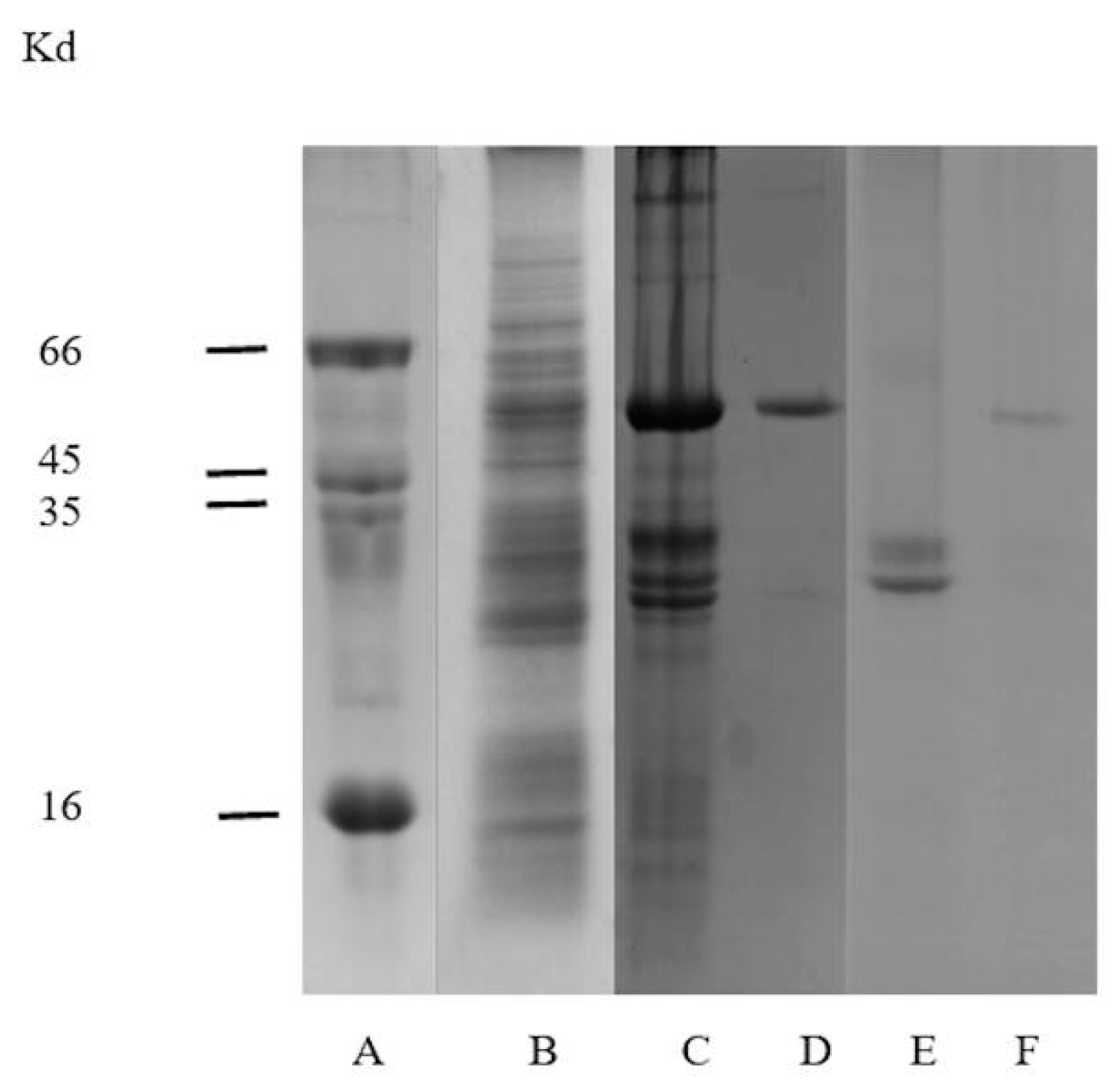
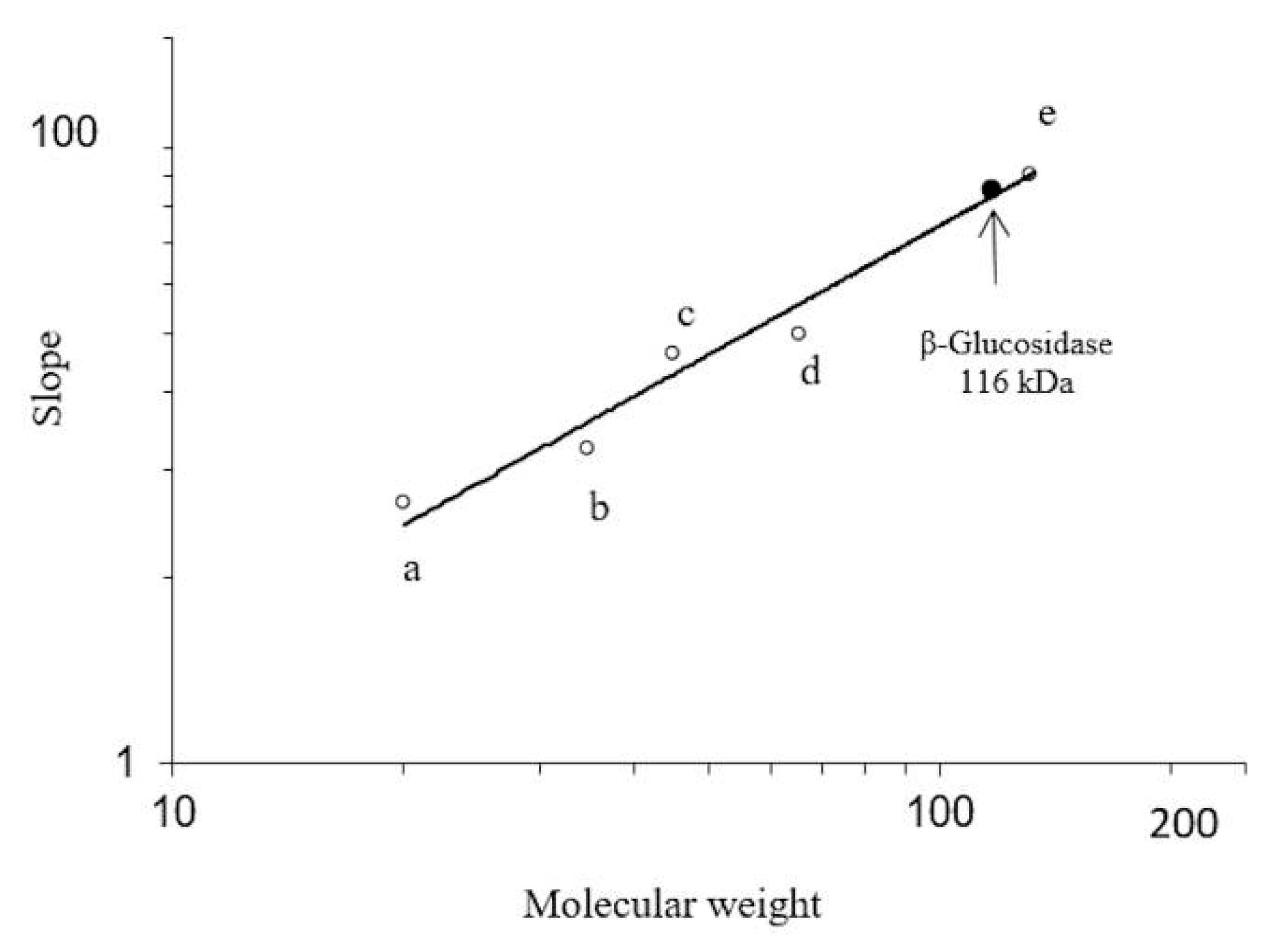
2.2. Electrophoretic Analysis
2.3. Peptide Analysis by Nano-LC-ESI-MS/MS
| Name | Mr a | Delta b | Peptide c | Start-End d Sequence |
|---|---|---|---|---|
| Sebg1 | 874.3643 | 0.0196 | MGFDAYR | 107-113- Csbg |
| Sebg2 | 865.446 | 0.0133 | FSIAWSR | 114-120- Csbg |
| Sebg3 | 982.4912 | −0.0026 | WFENFLK | 504-510- Csbg |
| Sebg4 | 1030.4430 | 0.0092 | DFSEVCFK | 184-191- Csbg |
| Sebg5 | 1087.5808 | −0.1337 | EAMQAGVRVK | 453-462- Csbg |
| Sebg6 | 1118.5648 | −0.0072 | FGLTYIDYK | 483-491- Csbg |
| Sebg7 | 1587.8297 | 0.9914 | NNLK | 492-495- Csbg |
2.4. Optimal Conditions for β-Glucosidase Activity


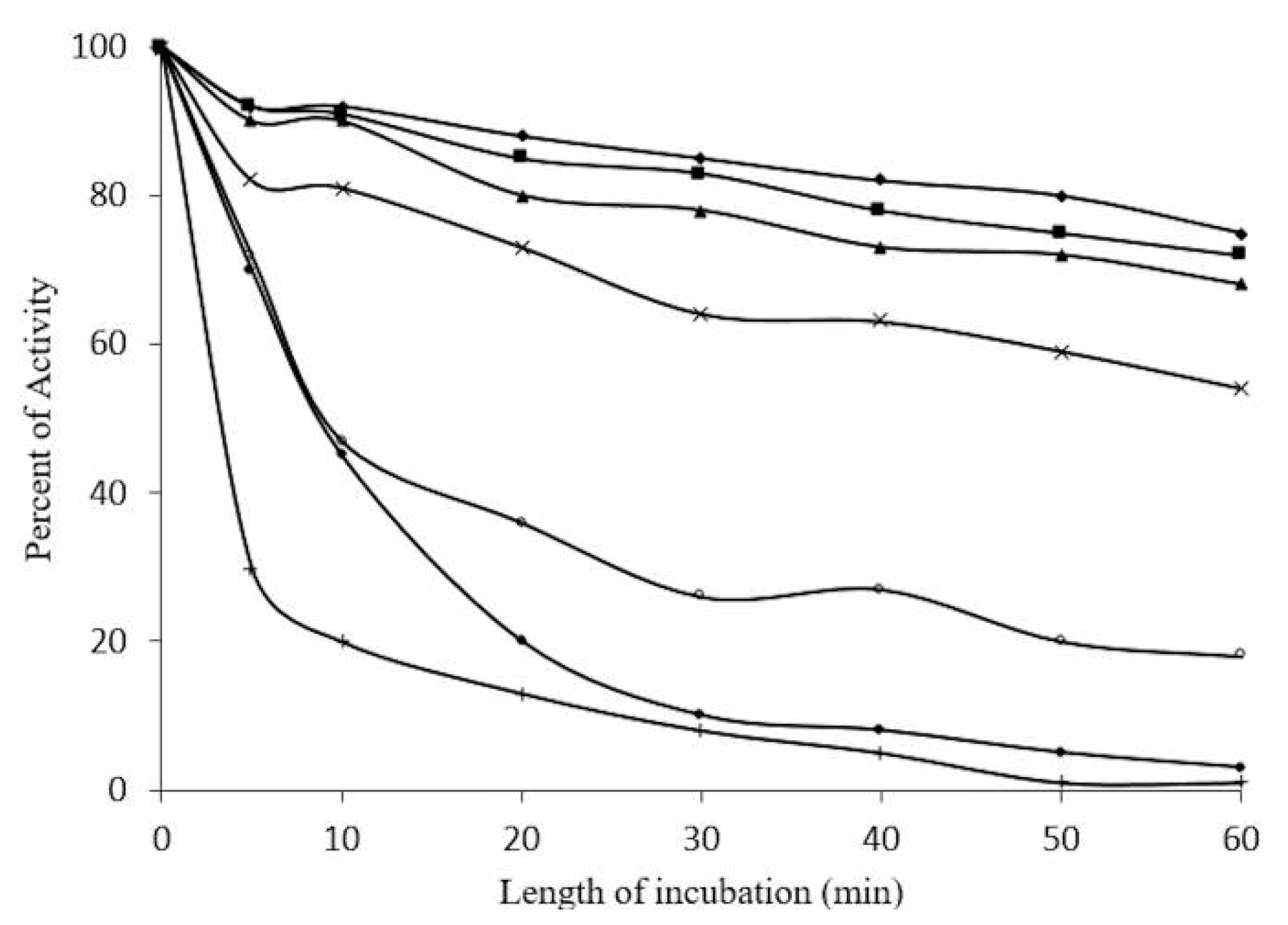


2.5. Concentration-Dependent stability of β-Glucosidase in S. edule
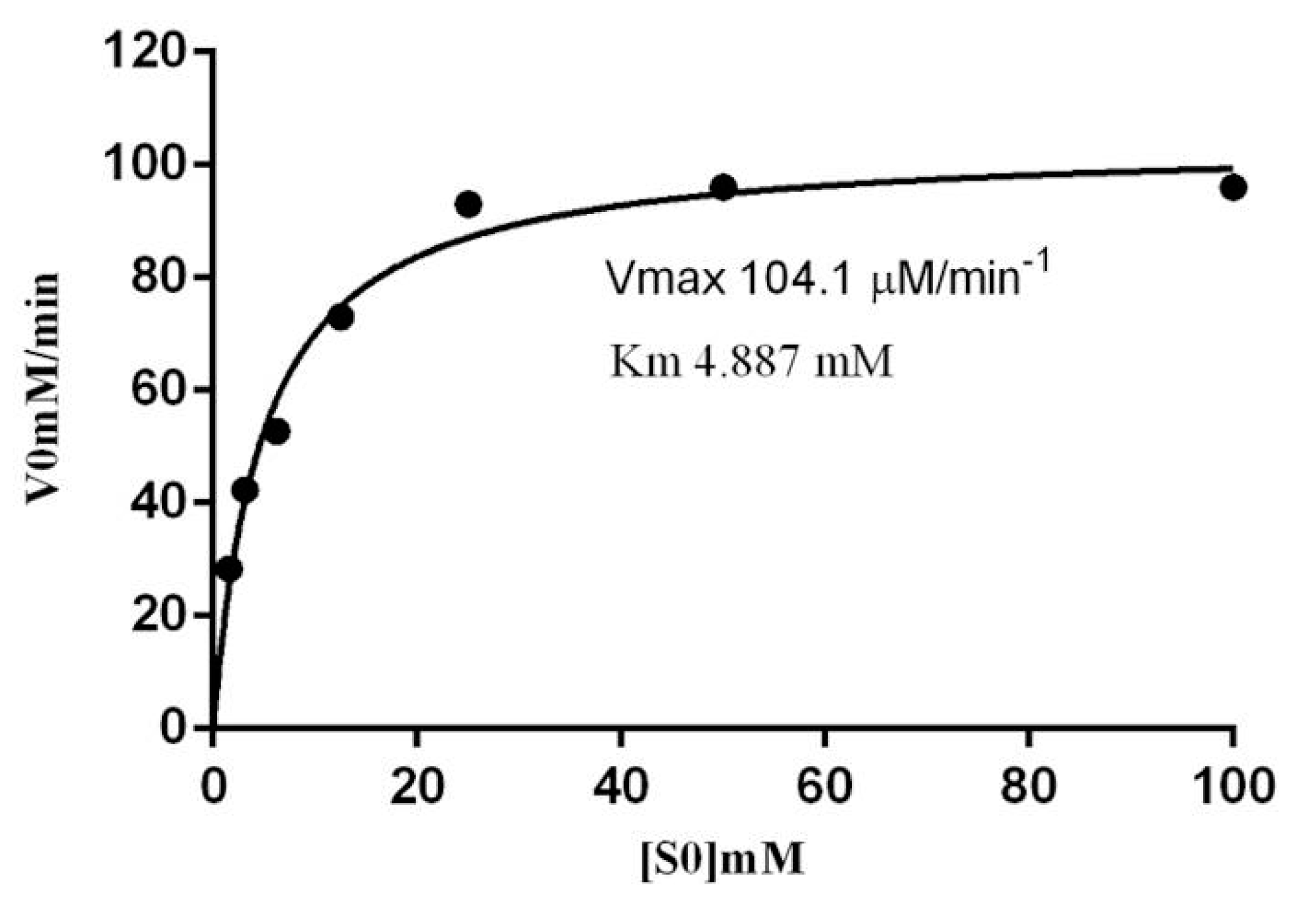
2.6. Kinetic Parameters of β-Glucosidase in S. edule
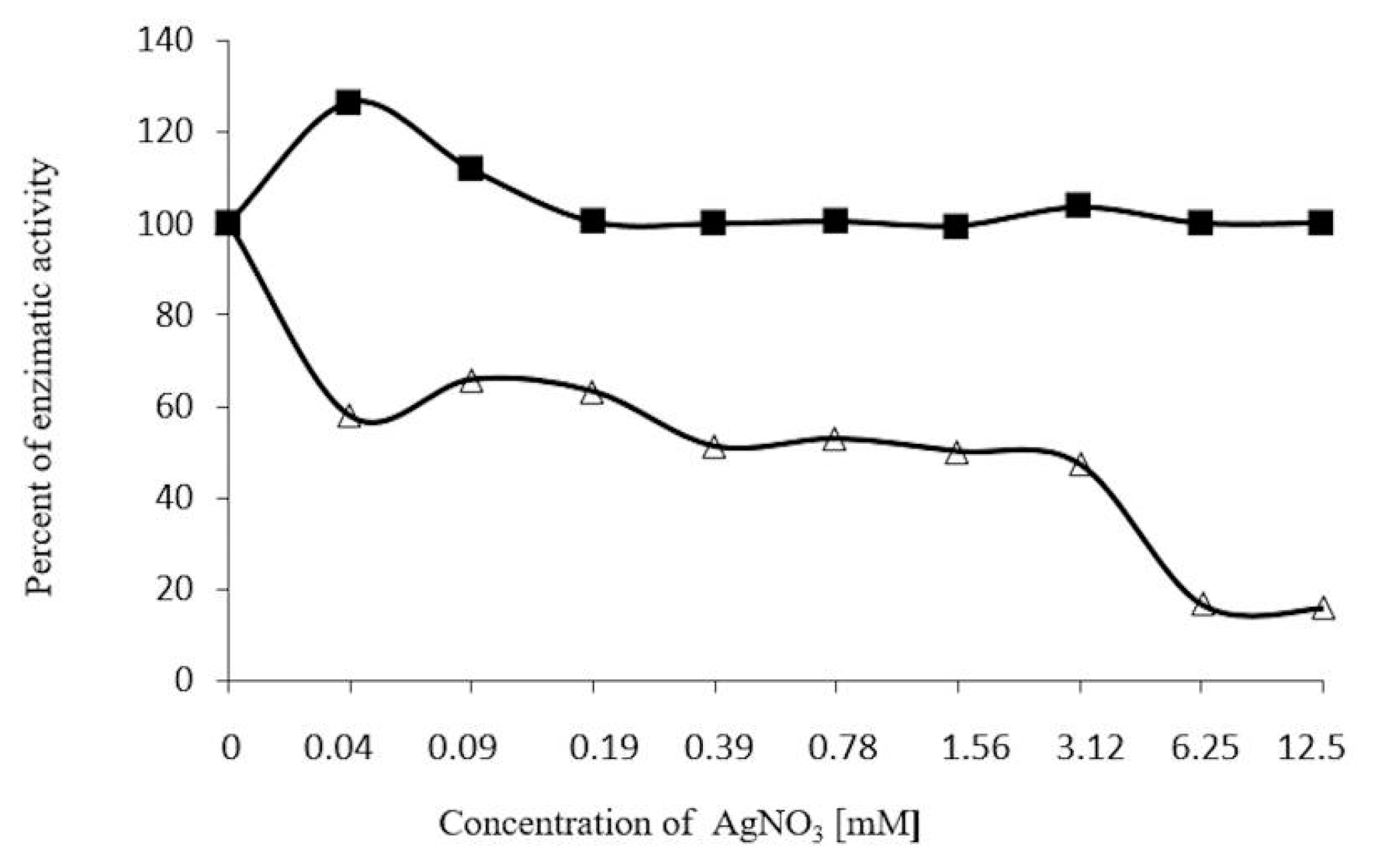
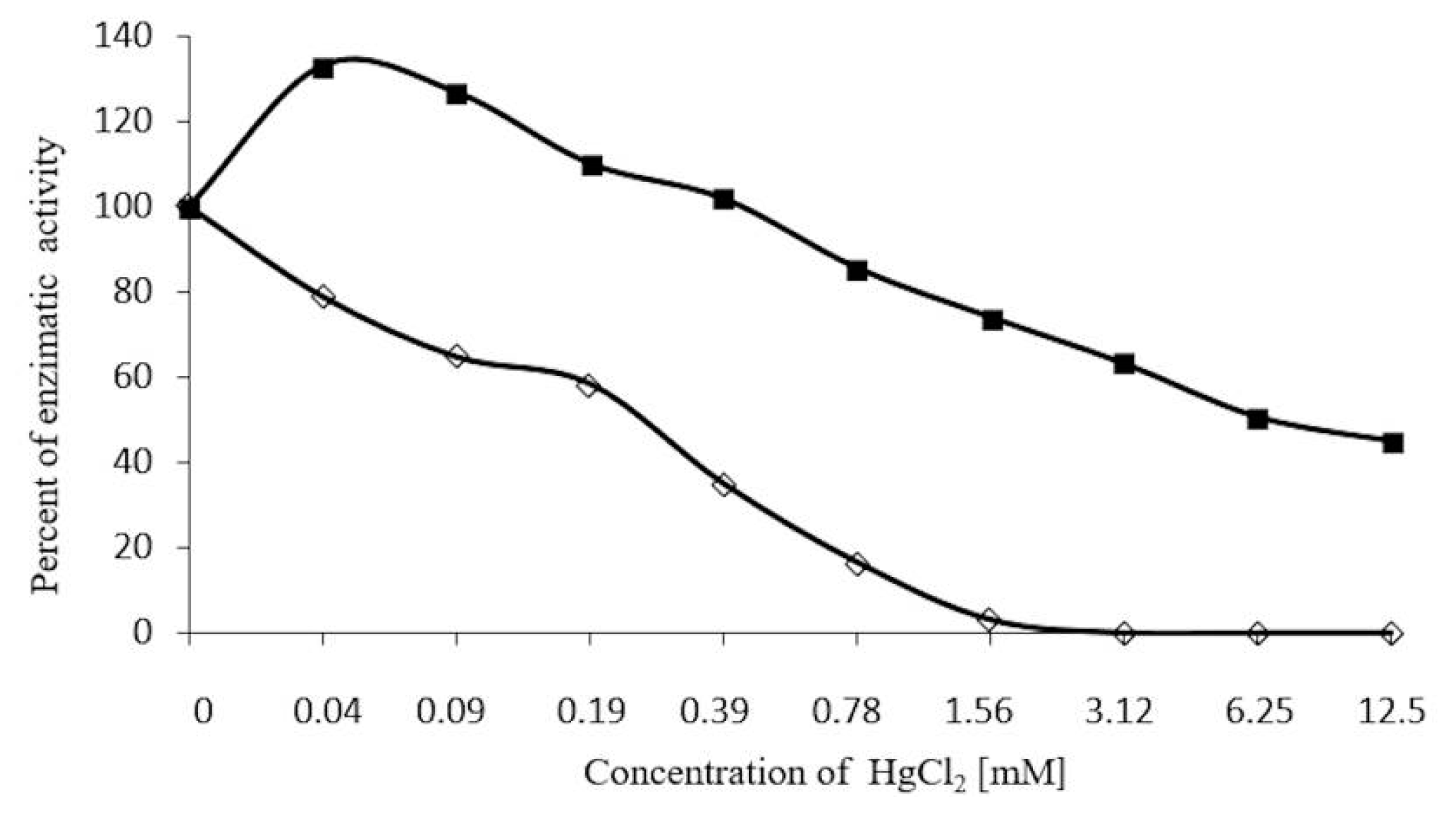
2.7. Inhibition Assays of β-Glucosidase in S. edule

3. Experimental Section
3.1. Protein Precipitation
3.2. Enzyme Purification
3.2.1. Cationic Interchange Chromatography
3.2.2. Size-Exclusion Chromatography
3.3. Determination of Molecular Mass
3.4. Peptide Mass Fingerprinting
3.5. Bioinformatic Analysis
3.6. Enzymatic Activity
3.7. Effects of pH and Temperature on Enzyme Activity
3.8. Effect of 2-ME, Ions, and Iodoacetamide on Enzyme Activity
3.9. Concentration-Dependent Stability of β-Glucosidase in S. edule
3.10. Inhibition Assays of β-Glucosidase in S. edule
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
References
- Henrissat, B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1991, 280, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Escamilla-Treviño, L.L.; Chen, W.; Card, M.L.; Shih, M.-C.H.; Cheng, C.-L.; Poulton, J.E. Arabidopsis thaliana β-glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides. Phytochemistry 2006, 67, 1651–1660. [Google Scholar] [CrossRef] [PubMed]
- Vanholme, R.; Morreel, K.; Ralph, J.; Boerjan, W. Lignin engineering. Curr. Opin. Plant Biol. 2008, 11, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, H.J.; Stålbrand, H.; Brumer, H. How the walls come tumbling down: Recent structural biochemistry of plant polysaccharide degradation. Curr. Opin. Plant Biol. 2008, 11, 338–348. [Google Scholar] [CrossRef] [PubMed]
- Lymar, E.S.; Li, B.; Renganathan, V. Purification and characterization of a cellulose-binding β-glucosidase from cellulose-degrading cultures of Phanerochaete chrysosporium. Appl. Environ. Microbiol. 1995, 61, 2976–2980. [Google Scholar] [PubMed]
- Igarashi, K.; Tani, T.; Rie, K.; Masahiro, S. Family 3 β-glucosidase from cellulose-degrading culture of the white-rot fungus Phanerochaete chrysosporium is a glucan 1,3-β-glucosidase. J. Biosci. Bioeng. 2003, 95, 572–576. [Google Scholar] [CrossRef]
- Morant, A.V.; Jørgenesen, K.; Jørgenesen, C.; Paquette, S.M.; Sánchez-Pérez, R.; Møller, B.L.; Bak, S. β-glucosidase as detonators of plant chemical defense. Phytochemistry 2008, 69, 1795–1813. [Google Scholar] [CrossRef] [PubMed]
- Morant, A.V.; Bjarnholt, N.; Kragh, M.E.; Kjaergaard, C.H.; Jørgenesen, K.; Paquette, S.M.; Piotrowski, M.; Imberty, A.; Olsen, C.E.; Møller, B.L.; et al. The β-glucosidase responsible for bioactivation of hydroxynitrile glucosides in Lotus japonicas. Plant Physiol. 2008, 147, 1072–1091. [Google Scholar] [CrossRef] [PubMed]
- Zagrobelny, M.; Bak, S.; Møller, B.L. Cyanogenesis in plants and arthropods. Phytochemistry 2008, 69, 1457–1468. [Google Scholar] [CrossRef] [PubMed]
- Pogorzelski, E; Wilkowska, A. Flavour enhancement through the enzymatic hydrolysis of glycosidic aroma precursors in juices and wine beverages: A review. Flavour Fragr. J. 2007, 22, 251–254. [Google Scholar]
- Orruño, E.; Apenten, R.-O.; Zabetakis, I. The role of β-glucosidase in biosintesis of 2,5-dimethyl-4-hydroxy-3(2H)-furanone in strawberry (Fragaria. X ananassa cv. Elsanta). Flavour Frag. J. 2001, 16, 81–84. [Google Scholar] [CrossRef]
- Sarry, J.-E.; Günata, Z. Plant and microbial glycoside hydrolases: Volatile release from glycosidic aroma precursors. Food Chem. 2004, 87, 509–521. [Google Scholar] [CrossRef]
- Sue, M.; Yamazaki, K.; Yajima, S.; Nomura, T.; Matsukawa, T.; Iwamura, H.; Miyamoto, T. Molecular and Structural Characterization of Hexameric β-d-Glucosidases in Wheat and Rye. Plant. Physiol. 2006, 141, 1237–1247. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Takahashi, S.; Watanabe, R.; Fukushima, Y.; Fujita, N.; Noguchi, A.; Yokoyama, R.; nishitani, K.; Nishino, T.; Nakayama, T. An isoflavone conjugate-hydrolyzing β-glucosidase from the roots of soybean (Glycine max) seedlings. J. Biol. Chem. 2006, 281, 30251–30259. [Google Scholar] [CrossRef] [PubMed]
- Ketuda Cairns, J.R.; Esen, A. β-glucosidase. Cell. Mol. Life Sci. 2010, 67, 3389–3405. [Google Scholar] [CrossRef] [PubMed]
- Gómes-Anduro, G.; Ceniceros-Ojeda, E.A.; Casados-Vázquez, L.E.; Bencivenni, C.; Sierra-Beltrán, A.; Murillo-Amador, B.; Tiessen, A. Genome-wide analysis of the β-glucosidase gene family in maize (Zea mays var. Var B 73). Plant Mol. Biol. 2011, 77, 159–183. [Google Scholar] [CrossRef] [PubMed]
- Poungbangpho, S.; Chuasuwan, S.; Chuphayak, C. Screening of β-d-galactosidase, β-d-glucosidase, and α-d-mannosidase in the seeds of cucurbitaceae. J. Naresuan Univ. 2000, 8, 56–67. [Google Scholar]
- Pessela, B.C.C.; Fuentes, M.; Mateo, C.; Munilla, R.; Carrascosa, A.V.; Fernandez-Lafuente, R.; Guisan, J.M. Purification and very strong reversible immobilization of large proteins on ionic exchangers by controlling the support and the immobilization conditions. Enzym. Microb. Technol. 2006, 39, 909–915. [Google Scholar] [CrossRef]
- Barbosa, O.; Ortiz, C.; Berenguer-Murcia, A.; Torres, R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Strategies for the one-step immobilization–purification of enzymes as industrial biocatalysts. Biotechnol. Adv. 2015, 33, 435–456. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.C.S.D.; Barbosa, O.; Ortiz, C.; Berenguer-Murcia, A.; Rodrigues, R.C.; Fernandez-Lafuente, R. Importance of the support properties for immobilization or purification of enzymes. ChemCatChem 2015, 7, 2413–2432. [Google Scholar] [CrossRef]
- Xiao, Z.; Zhang, X.; Gregg, D.J.; Saddler, J.N. Effects of sugar inhibition on cellulases and β-glucosidase during enzymatic hydrolysis of softwood substrates. Appl. Biochem. Biotechnol. 2004, 113, 1115–1126. [Google Scholar] [CrossRef]
- Liu, L.; Deseo, M.A.; Morris, C.; Winter, K.M.; Leach, D.N. Investigation of α-glucosidase inhibitory activity of wheat bran and germ. Food Chem. 2011, 126, 553–561. [Google Scholar] [CrossRef]
- Esen, A. Purification and partial characterization of maize (Zea mays) β-glucosidase. Plant Physiol. 1992, 98, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Cicek, M.; Esen, A. Structure and expression of a dhurrinase (β-glucosidase) from sorghum. Plant Physiol. 1998, 116, 1469–1478. [Google Scholar] [CrossRef] [PubMed]
- Leah, R.; Kigel, J.; Svendsen, I.; Mundy, J. Biochemical and molecular characterization of a barley seed β-glucosidase. J. Biol. Chem. 1995, 270, 15789–15797. [Google Scholar] [CrossRef] [PubMed]
- Hrmova, M.; Harvey, A.J.; Wang, J.; Shirley, N.J.; Jones, G.P.; Stone, B.A.; Høj, P.B.; Fincher, G.B. Barley β-d-glucan exohydrolases with β-d-glucosidase activity. J. Biol. Chem. 1996, 271, 5277–5286. [Google Scholar] [PubMed]
- Hrmova, M.; MacGregor, E.A.; Biely, P.; Stewart, R.J.; Fincher, G.B. Substrate binding and catalytic mechanism of a barley β-d-glucosidase/(1,4)-β-d-glucan exohydrolase. J. Biol. Chem. 1998, 273, 11134–11143. [Google Scholar] [CrossRef] [PubMed]
- Opassiri, R.; Ketudat Cairns, J.R.; Akiyama, T.; Wara-Aswapati, O.; Svasti, J.; Esen, A. Characterization of a rice β-glucosidase highly expressed in flower and germinating shoot. Plant Sci. 2003, 165, 627–638. [Google Scholar] [CrossRef]
- Akiyama, T.; Kaku, H.; Shibuya, N. A cell wall-bound beta-glucosidase from germinated rice: purification and properties. Phytochemistry 1998, 48, 49–54. [Google Scholar] [CrossRef]
- Czjzek, M.; Cicek, M.; Zamboni, V.; Bevan, D.R.; Henrissat, B.; Esen, A. The Mechanism of substrate (aglycone) specificity in β-glucosidase is revealed by crystal structures of mutant maize β-glucosidase-DIMBOA, DIMBOA-Glc and-dhurrin complexes. Proc. Natl. Acad. Sci. USA 2000, 97, 13555–13560. [Google Scholar] [CrossRef] [PubMed]
- Czjzek, M.; Cicek, M.; Zamboni, V.; Burmeister, W.P.; Bevan, D.R.; Henrissat, B.; Esen, A. Crystal structure of a monocotyledon (maize ZMGlu1) β-glucosidase and a model of its complex with p-nitrophenyl β-d-thioglucoside. Biochem. J. 2001, 354, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Alpuche, J.; Pereyra, A.; Mendoza-Hernández, G.; Agundis, C.; Rosas, C.; Zenteno, E. Purification and partial characterization of agglutinin from Octopus maya serum. Comp. Biochem. Phys. B 2010, 156, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Findlay, J.B.; Pappin, D.J.; Keen, J. Automated solid-phase microsequencing. In Protein Sequencing: A Practical Approach; Findlay, J.B., Geisow, M.J., Eds.; IRL Publishers: Oxford, UK, 1989; pp. 69–84. [Google Scholar]
- Esen, A. β-Glucosidases. In Handbook of Food Enzymology; Whitaker, J.R., Voragen, A.G.J., Wong, D.W.S., Eds.; Marcel Dekker Inc.: New York, NY, USA, 2003; pp. 791–804. [Google Scholar]
- Smythe, C.V. The reaction of iodoacetate and of iodoacetamide with variuos sulfhydryl groups, with urease, and with yeast preparations. J. Biol. Chem. 1936, 114, 601–612. [Google Scholar]
- Anfinsen, C.B. Principles that govern folding of protein chains. Science 1973, 181, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Krajewska, B. Mono-(Ag, Hg) and di-(Cu, Hg) valent metal ions effects on the activity of jack bean urease. Probing the modest of metal binding to the enzyme. J. Enzyme Inhib. Med. Chem. 2008, 4, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Waku, K.; Nakazawa, N.Y. Toxic effect of several mercury compounds on SH- and non-SH enzymes. Toxicol. Lett. 1979, 4, 49–55. [Google Scholar]
- Yu, H.Y.; Kittur, F.S.; Bevan, D.R.; Esen, A. Determination of β-glucosidase aggregating factor (BGAF) binding and polymerization regions on the maize β-glucosidase isozyme Glu1. Phytochemistry 2009, 70, 1355–1365. [Google Scholar] [CrossRef] [PubMed]
- Vozari-Hampe, M.M.; Viegas, C.; Saucedo, C.; Rosseto, S.; Manica, G.G.; Hampe, O.G. A lectin from Sechium edule fruit exudates. Phytochemistry 1992, 31, 1477–1480. [Google Scholar] [CrossRef]
- Rosales-Calderon, O.; Trajano, H.L.; Duff, S.J.B. Stability of comercial glucanase and β-glucosidase preparations under hydrolysis conditions. Peer J. 2014, 2, e402. [Google Scholar] [CrossRef] [PubMed]
- Parry, N.J.; Beever, D.E.; Owen, E.; Vandenberghe, I.; van Beeumen, J.; Bhat, M.K. Biochemical characterization and mechanism of action of a thermostable beta-glucosidase purified from Thermoascus aurantiacus. Biochem. J. 2001, 353, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, R.C.; Ortiz, C.; Berenguer-Murcia, A.; Torres, R.; Fernández-Lafuente, R. Modifying enzyme activity and selectivity by immobilization. Chem. Soc. Rev. 2013, 42, 6290–6307. [Google Scholar] [CrossRef] [PubMed]
- Mateo, C.; Palomo, J.M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Improvement of enzyme activity, stability and selectivity via immobilization techniques. Enzyme Microb. Technol. 2007, 40, 1451–1463. [Google Scholar] [CrossRef]
- Guzik, U.; Hupert-Kocurek, K.; Wojcieszyńska, D. Immobilization as a strategy for improving enzyme propierties-application to oxidoreductases. Molecules 2014, 19, 8995–9018. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhao, S.; Yin, P.; Yan, L.; Han, J.; Shi, L.; Zhou, X.; Liu, Y.; Ma, C. α-Glucosidase inhibitory activity of polyphenols from the burs of Castanea mollissima Blume. Molecules 2014, 19, 8373–8386. [Google Scholar] [CrossRef] [PubMed]
- Mateo, C.; Palomo, J.M.; Fuentes, M.; Betancor, L.; Grazu, V.; López-Gallego, F.; Pessela, B.C.C.; Hidalgo, A.; Fernández-Lorente, G.; Fernández-Lafuente, R.; et al. Glyoxyl agarose: A full inert and hydrophilic support for immobilization and high stabilization of proteins. Enzyme Microb. Technol. 2006, 39, 274–280. [Google Scholar] [CrossRef]
- Mateo, C.; Fernandez-Lorente, G.; Pessela, B.C.C.; Vian, A.; Carrascosa, A.V.; García, J.L.; Fernandez—Lafuente, R; Guisan, J.M. Affinity chromatography of polyhistidine tagged enzymes new dextran-coated immobilized metal ion affinity chromatography matrices for prevetion of undesired multipoint adsorptions. J. Chromatogr. A. 2001, 915, 97–106. [Google Scholar] [CrossRef]
- Fuentes, M.; Pessela, B.C.C.; Maquiese, J.V.; Ortiz, C.; Segura, R.L.; Palomo, J.M.; Abian, O.; Torres, R.; Mateo, C.; Fernández-Lafuente, R.; Guisán, J.M. Reversible and strong immobilization of proteins by ionic exchange on supports coated with sulfate-dextran. Biotechnol. Prog. 2004, 20, 1134–1139. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for quantitation of micrograma quantities of proteins utilizing the principles of protein dye-binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleave of structural proteins during the assembly of the head of bacteriophase T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Hedrick, J.L.; Smith, A.J. Size and charge isomer separation and estimation of molecular weights of proteins by disc gel electrophoresis. Arch. Biochem. Biophys. 1968, 126, 155–164. [Google Scholar] [CrossRef]
- Goujon, M.; McWilliam, H.; Li, W.; Squizzato, S.; Paern, J.; Lopez, R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acid Res. 2010, 38, W695–W699. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the β-Glucosidase from chayote (Sechium edule). are available from the authors.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mateos, S.E.; Cervantes, C.A.M.; Zenteno, E.; Slomianny, M.-C.; Alpuche, J.; Hernández-Cruz, P.; Martínez-Cruz, R.; Canseco, M.D.S.P.; Pérez-Campos, E.; Rubio, M.S.; et al. Purification and Partial Characterization of β-Glucosidase in Chayote (Sechium edule). Molecules 2015, 20, 19372-19392. https://doi.org/10.3390/molecules201019372
Mateos SE, Cervantes CAM, Zenteno E, Slomianny M-C, Alpuche J, Hernández-Cruz P, Martínez-Cruz R, Canseco MDSP, Pérez-Campos E, Rubio MS, et al. Purification and Partial Characterization of β-Glucosidase in Chayote (Sechium edule). Molecules. 2015; 20(10):19372-19392. https://doi.org/10.3390/molecules201019372
Chicago/Turabian StyleMateos, Sergio Espíndola, Carlos Alberto Matías Cervantes, Edgar Zenteno, Marie-Christine Slomianny, Juan Alpuche, Pedro Hernández-Cruz, Ruth Martínez-Cruz, Maria Del Socorro Pina Canseco, Eduardo Pérez-Campos, Manuel Sánchez Rubio, and et al. 2015. "Purification and Partial Characterization of β-Glucosidase in Chayote (Sechium edule)" Molecules 20, no. 10: 19372-19392. https://doi.org/10.3390/molecules201019372








