EDTA a Novel Inducer of Pisatin, a Phytoalexin Indicator of the Non-Host Resistance in Peas
Abstract
:1. Introduction

Molecular and Cytological Observations of the Pea Immune Response
2. Results and Discussion
2.1. Phytoalexin Induction by EDTA
| Treatment a | Conc. EDTA mM | Pisatin μg/g frs.wt. |
|---|---|---|
| Water | 0 | 0.0 |
| EDTA | 250 | 0.0 |
| EDTA | 125 | 12.4 ± 9.7 |
| EDTA | 62.5 | 62.2 ± 22.2 |
| EDTA | 31.2 | 194.2 ± 53.7 |
| EDTA | 15.6 | 136.2 ± 5.5 |
| EDTA | 7.8 | 85.8 ± 19.3 |
| EDTA | 3.9 | 3.3 ± 2.9 |
| EDTA | 1.9 | 0.6 ± 0.5 |
| EDTA | 0.9 | 0.0 ± 0.0 |
| Water + Fsph spores | 0.0 | 221.5 ± 67.9 |
| EDTA + Fsph spores | 250 | 1.7 ± 1.7 |
| EDTA + Fsph spores | 125 | 15.1 ± 2.1 |
| EDTA + Fsph spores | 62.5 | 86.3 ± 48.3 |
| EDTA + Fsph spores | 31.2 | 292.2 ± 115.2 |
| EDTA + Fsph spores | 15.6 | 428.9 ± 29.5 |
| EDTA + Fsph spores | 7.8 | 474.8 ± 16.3 |
| EDTA + Fsph spores | 3.9 | 353.6 ± 3.1 |
| EDTA + Fsph spores | 1.9 | 376.9 ± 39.7 |
| EDTA + Fsph spores | 0.9 | 258.0 ± 53.0 |
2.2. Effect of EDTA on the Defense Response of Pea; Defense Gene Activation
| Target | Genbank # | Real-time F primer | Real-time R primer |
|---|---|---|---|
| Pea Ubiquitin | L881142 | GGCTAAGATACAGGACAAGGAG | AACGAAGGACAAGATGAAGGG |
| Pea Actin (Pea-ACT) | U81046 | CACAATTGGCGCTGAAAGATT | GATCATCGATGGCTGGAACA |
| DRR206 | U11716 | CTTGGCTTAGTTTCACATTTGTTCTT | GGGTCAGCTCCAGCAAAAGTAA |
| DRR230 (defensin) | L01579 | TGTGGTGACAGAGGCAAACAC | TCGTGAAGCATACTCCCCTGTA |
| PR10 (AKA DRR49) | U31669 | GATCTCATTCGAGGCTAAACTGTCT | CACACTCAGCTTTGCAATGGA |
| PR1b | AJ586324.1 | AACTCATGTGCTGCTGGTTATCA | AACCGAATTGCGCCAAAC |
2.3. Direct Effect of EDTA on Fungal DNase

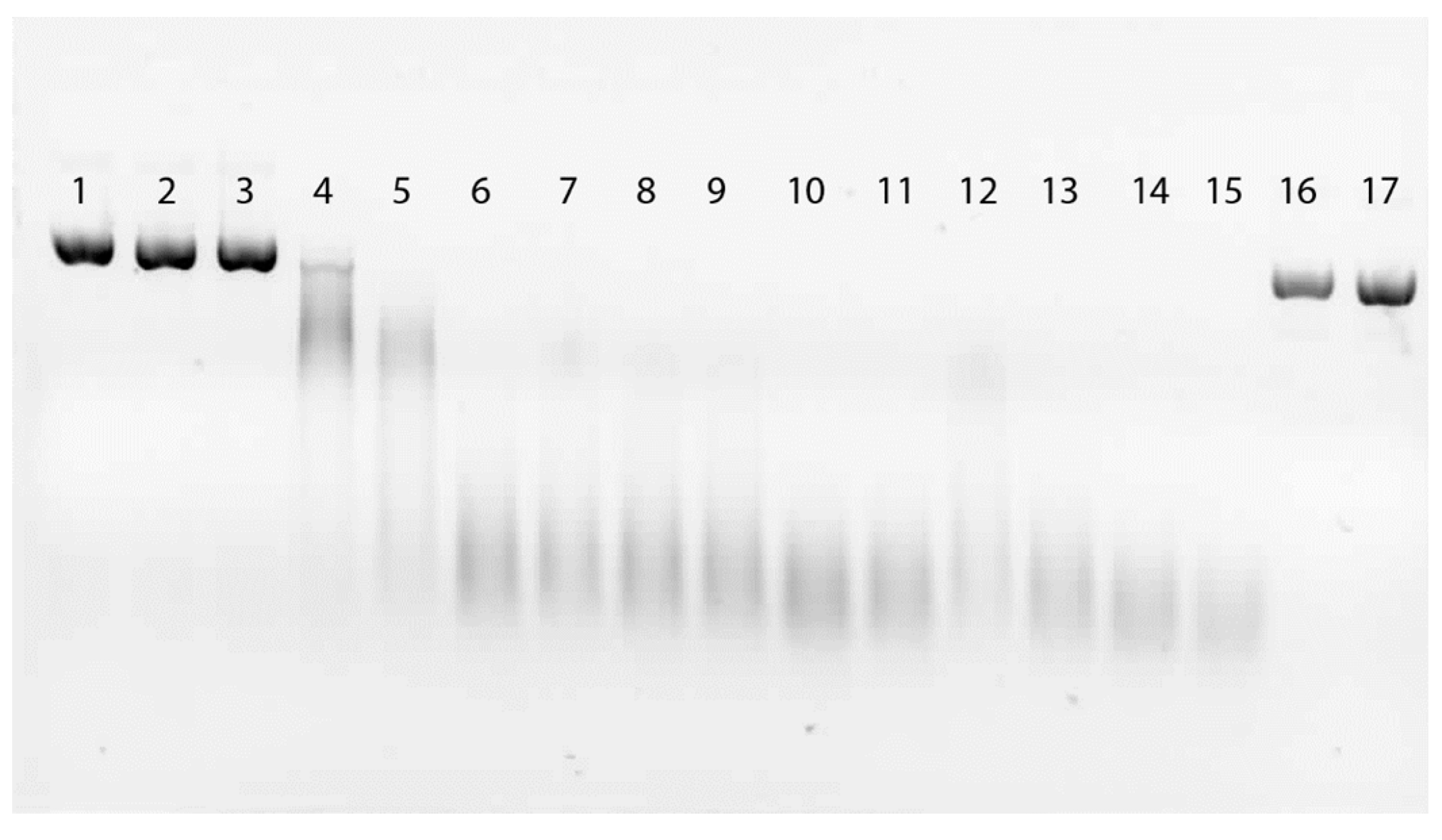
2.4. DNA Damage by EDTA

2.5. Nonhost Resistance Induced by EDTA
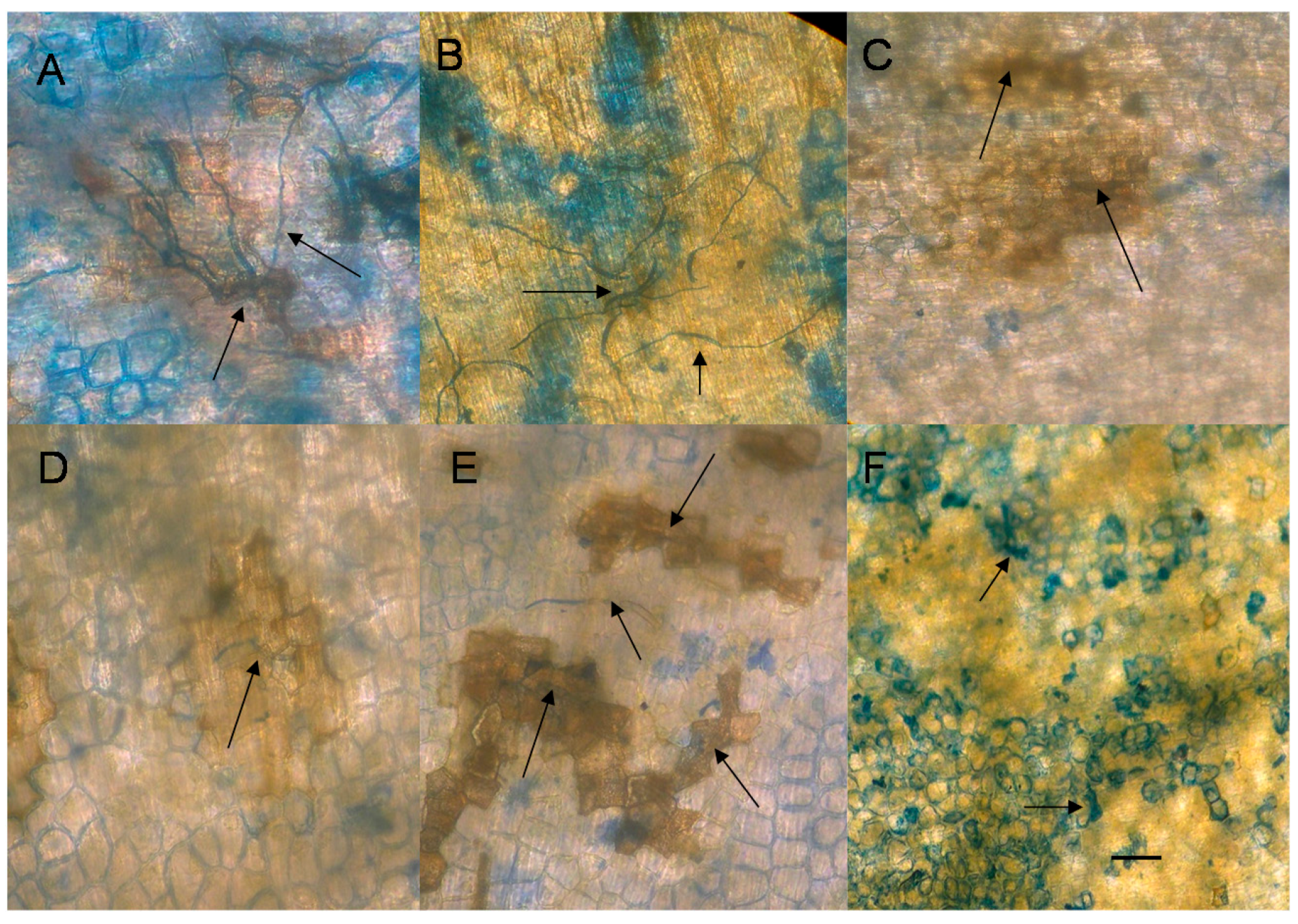
2.6. Cytological Effect of EDTA on Pea Nuclear Condition

3. Experimental Section
3.1. Plant and Pathogens
3.2. EDTA Treatment and Pisatin Quantization
3.3. Activation of PR Genes
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hadwiger, L.A. Pea-Fusarium solani interactions: Contributions of a system toward understanding disease resistance. Phytopathology 2008, 98, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Klosterman, J.; Chen, J.; Choi, J.J.; Chinn, E.E.; Hadwiger, L.A. Characterization of a 20 kDa DNase elicitor from Fusarium solani f. sp. phaseoli and its expression at the onset of induced resistance in Pisum sativum. Mol. Plant Pathol. 2001, 2, 147–158. [Google Scholar]
- Gerhold, D.L.; Pettinger, A.J.; Hadwiger, L.A. Characterization of a plant stimulated nuclease from Fusarium solani. Physiol. Mol. Plant Pathol. 1933, 43, 33–46. [Google Scholar] [CrossRef]
- Hartney, S.; Carson, J.; Hadwiger, L.A. The use of chemical genomics to detect functional systems affecting the non-host disease resistance of pea to Fusarium solani f. sp. phaseoli. Plant Sci. 2007, 172, 45–56. [Google Scholar] [CrossRef]
- Flora, S.J.S.; Pachauri, V. Chelation in metal intoxication. Int. J. Environ. Res. Public Health 2010, 7, 2745–2788. [Google Scholar] [CrossRef] [PubMed]
- Drouin, R.; Gao, S.; Holmquist, G.P. Technologies for Detection of DNA Damage and Mutations; Pfeifer, G.P., Ed.; Plenum Press: New York, NY, USA, 1996; pp. 37–43. [Google Scholar]
- Isaac, J.; Hartney, S.L.; Druffel, K.; Hadwiger, L.A. The non-host disease resistance response in peas; alterations in phosphorylation and ubiquitination of HMG A and histones. H2A/H2B. Plant Sci. 2009, 177, 439–449. [Google Scholar] [CrossRef]
- Pilet-Nayel, M.L.; Muehlbauer, F.J.; McGee, R.J.; Kraft, J.M.; Baranger, A.; Coyne, C.J. Quantitative trait loci for partial resistance to Aphanomyces root rot in pea. Theor. Appl. Genet. 2002, 106, 28–39. [Google Scholar] [PubMed]
- Prioul-Gervais, S.; Deniot, G.; Recevur, E.-M.; Frankewitz, A.; Fourmann, M.; Rameau, C.; Pilet-Nayel, M.-L.; Baranger, A. Candidate genes for quantitative resistance to Mycosphaerella pinodes in peas (Pisum sativum L.). Theor. Appl. Genet. 2007, 114, 971–984. [Google Scholar] [CrossRef] [PubMed]
- Hadwiger, L.A. Localization predictions for gene products involved in non-host resistance responses in a model plant/fungal pathogen interaction. Plant Sci. 2009, 177, 257–265. [Google Scholar] [CrossRef]
- DiCenzo, G.L.; VanEtten, H.D. Studies on the late steps of (+) pisatin biosynthesis: Evidence for (−) enatiomeric intermediates. Phytochemistry 2006, 67, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Hadwiger, L.A.; Polashock, J. Fungal mitochondrial DNases: Effectors with the potential to activate plant defenses in nonhost resistance. Phytopathology 2013, 103, 81–90. [Google Scholar] [CrossRef]
- Chiang, C.C.; Hadwiger, L.A. The Fusarium solani-induced expression of a pea gene family encoding cysteine content proteins. MPMI 1991, 4, 324–331. [Google Scholar] [CrossRef]
- Almeida, M.S.; Cabral, K.M.; Zingali, R.B.; Kurtenbach, E. Characterization of two novel defense peptides from pea (Pisum sativum) seeds. Arch. Biochem. Biophys. 2006, 378, 278–286. [Google Scholar] [CrossRef]
- Allaire, B.S.; Hadwiger, L.A. Immunogold localization of a disease resistance response protein in Pisum sativum endocarp cells. Physiol. Mol. Plant Pathol. 1994, 44, 9–17. [Google Scholar] [CrossRef]
- Chang, M.M.; Chiang, C.C.; Martin, M.W.; Hadwiger, L.A. Expression of a pea disease resistance response protein in potatoes. Am. Potato J. 1993, 70, 635–647. [Google Scholar] [CrossRef]
- Yu, D.; Chen, C.; Zhixiang, C. Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 2001, 13, 1527–1539. [Google Scholar] [CrossRef] [PubMed]
- Maier, F.; Zwicker, S.; Huckelhoven, A.; Meissner, M.; Funk, J.; Pfitznener, S.J.; Pfitzner, U.M. Nonexpressor of Pathogenesis-Related Proteins 1 (NPR1) and some NPR1-related proteins are sensitive to salicylic acid. Mol. Plant Pathol. 2011, 12, 73–91. [Google Scholar] [CrossRef] [PubMed]
- Barr, R.; Troxel, K.S.; Crane, F.L. EGTA, a calcium chelator, inhibits electron transport in photosystem II of spinach chloroplasts at two different sites. Biochem. Biophys. Res. Commun. 1980, 92, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Weake, V.M.; Workman, J.L. Histone ubiquitination triggering gene activity. Mol. Cell 2008, 29, 653–663. [Google Scholar] [CrossRef] [PubMed]
- Adkins, M.W.; Tyler, J.K. Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Mol. Cell 2006, 21, 405–416. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Wu, H.; Wu, Y.; Wang, C.; Zhang, H.; Shi, X.; Yang, J. High molecular weight Hyaluronan decreases oxidative DNA damage induced by EDTA in human coryneal epithelial cells. Eye 2012, 26, 1012–1020. [Google Scholar] [CrossRef] [PubMed]
- Schochau, M.E.; Hadwiger, L.A. Regulation of gene expression by actinomycin D and other compounds which change the conformation of DNA. Arch. Biochem. Biophys. 1969, 134, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Hadwiger, L.A.; Jafri, A.; von Broembsen, S.; Eddy, R. Mode of pisatin induction. Increased template activity and dye-binding capacity of chromatin isolated from polypeptide-treated pea pods. Plant Physiol. 1974, 53, 52–63. [Google Scholar]
- Choi, J.J.; Klosterman, S.J.; Hadwiger, L.A. A comparison of the effects of DNA-damaging agents and biotic elicitors on the induction of plant defense genes, nuclear distortion and cell death. Plant Physiol. 2001, 125, 752–762. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples described in this publication are available upon request in a timely manner for noncommercial research purposes.
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hadwiger, L.A.; Tanaka, K. EDTA a Novel Inducer of Pisatin, a Phytoalexin Indicator of the Non-Host Resistance in Peas. Molecules 2015, 20, 24-34. https://doi.org/10.3390/molecules20010024
Hadwiger LA, Tanaka K. EDTA a Novel Inducer of Pisatin, a Phytoalexin Indicator of the Non-Host Resistance in Peas. Molecules. 2015; 20(1):24-34. https://doi.org/10.3390/molecules20010024
Chicago/Turabian StyleHadwiger, Lee A., and Kiwamu Tanaka. 2015. "EDTA a Novel Inducer of Pisatin, a Phytoalexin Indicator of the Non-Host Resistance in Peas" Molecules 20, no. 1: 24-34. https://doi.org/10.3390/molecules20010024






