Catalytic Behavior of Lipase Immobilized onto Congo Red and PEG-Decorated Particles
Abstract
:1. Introduction
2. Results and Discussion
2.1. Characteristics of PS/PEG and PS/PEG/CR Particles
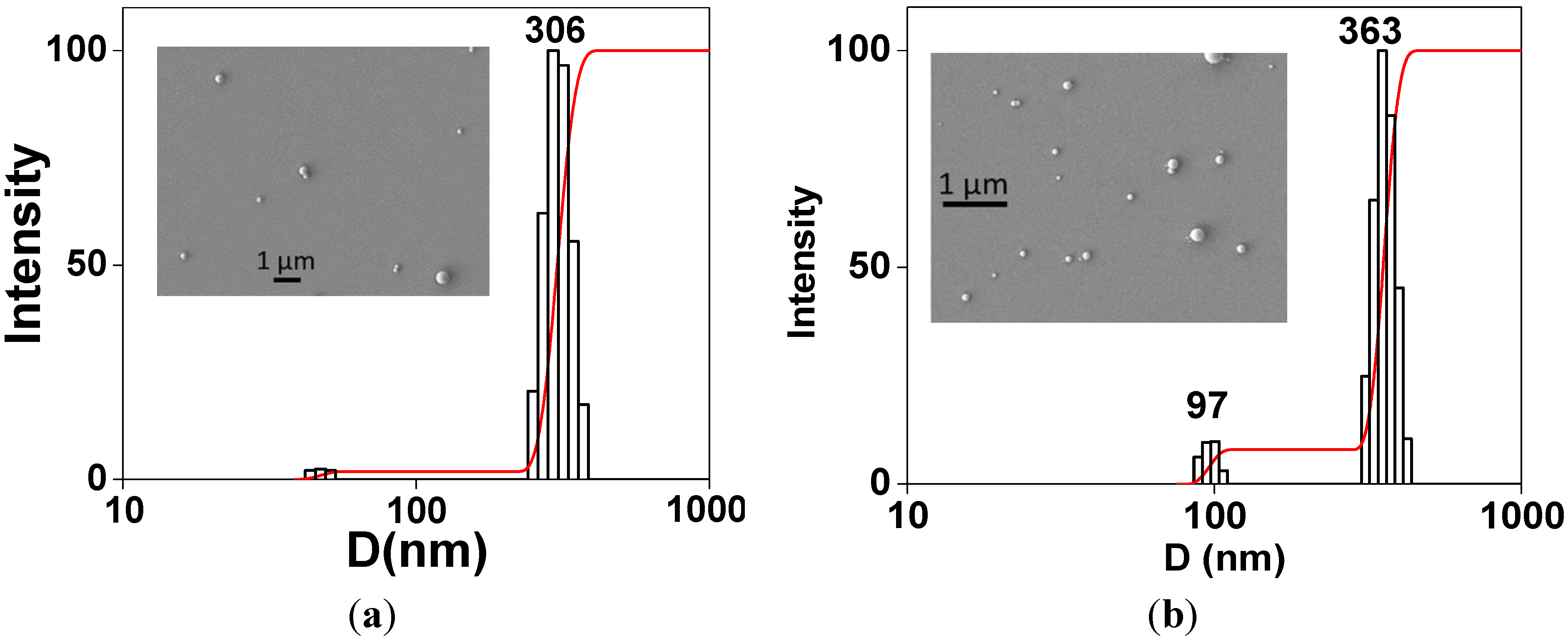
| D (nm) | ζ (mV) | P | |
|---|---|---|---|
| PS/PEG | 286 ± 15 | −(50 ± 5) | 0.17 ± 0.01 |
| PS/PEG/CR | 290 ± 19 | −(36 ± 5) | 0.21 ± 0.02 |
| PS/PEG/lipase (a) | 380 ± 20 | −(32 ± 4) | 0.20 ± 0.03 |
| PS/PEG/CR/lipase (b) | 405 ± 11 | −(25 ± 2) | 0.27 ± 0.05 |
2.2. Adsorption Behavior of Lipase onto PS/PEG or PS/PEG/CR Particles

2.3. Enzymatic Activity of Free and Adsorbed Lipase
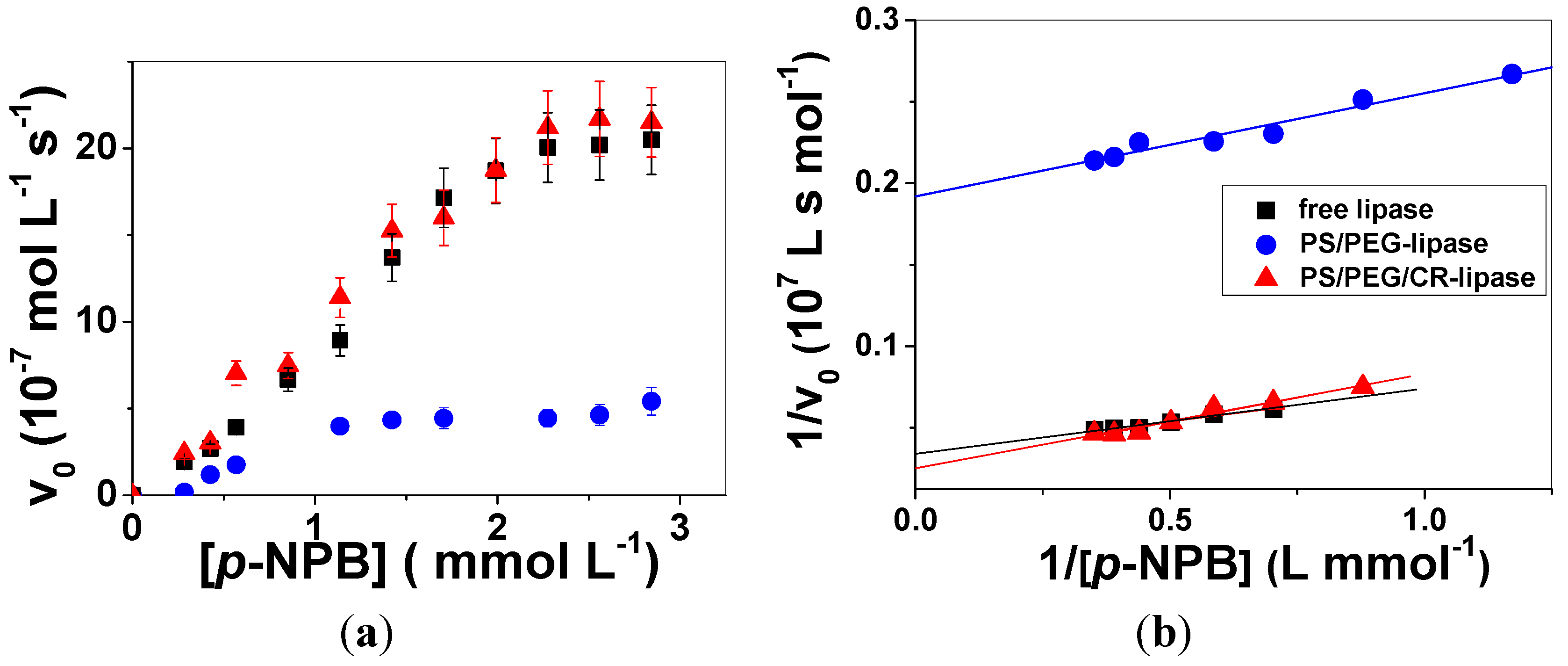
| System | Vmax (10−7 mol∙L−1∙s−1) | KM (10−3 mol∙L−1) | kcat (s−1) | ε (L s−1∙mol−1) |
|---|---|---|---|---|
| Free lipase | 30.3 ± 0.5 | 1.25 ± 0.08 | 7.5 ± 0.8 | 6000 ± 691 |
| PS/PEG/CR-Lipase | 46 ± 1 | 2.9 ± 0.5 | 11 ± 1 | 3958 ± 344 |
| PS/PEG-Lipase | 5.2 ± 0.5 | 0.32 ± 0.04 | 1.3 ± 0.1 | 4053 ± 678 |
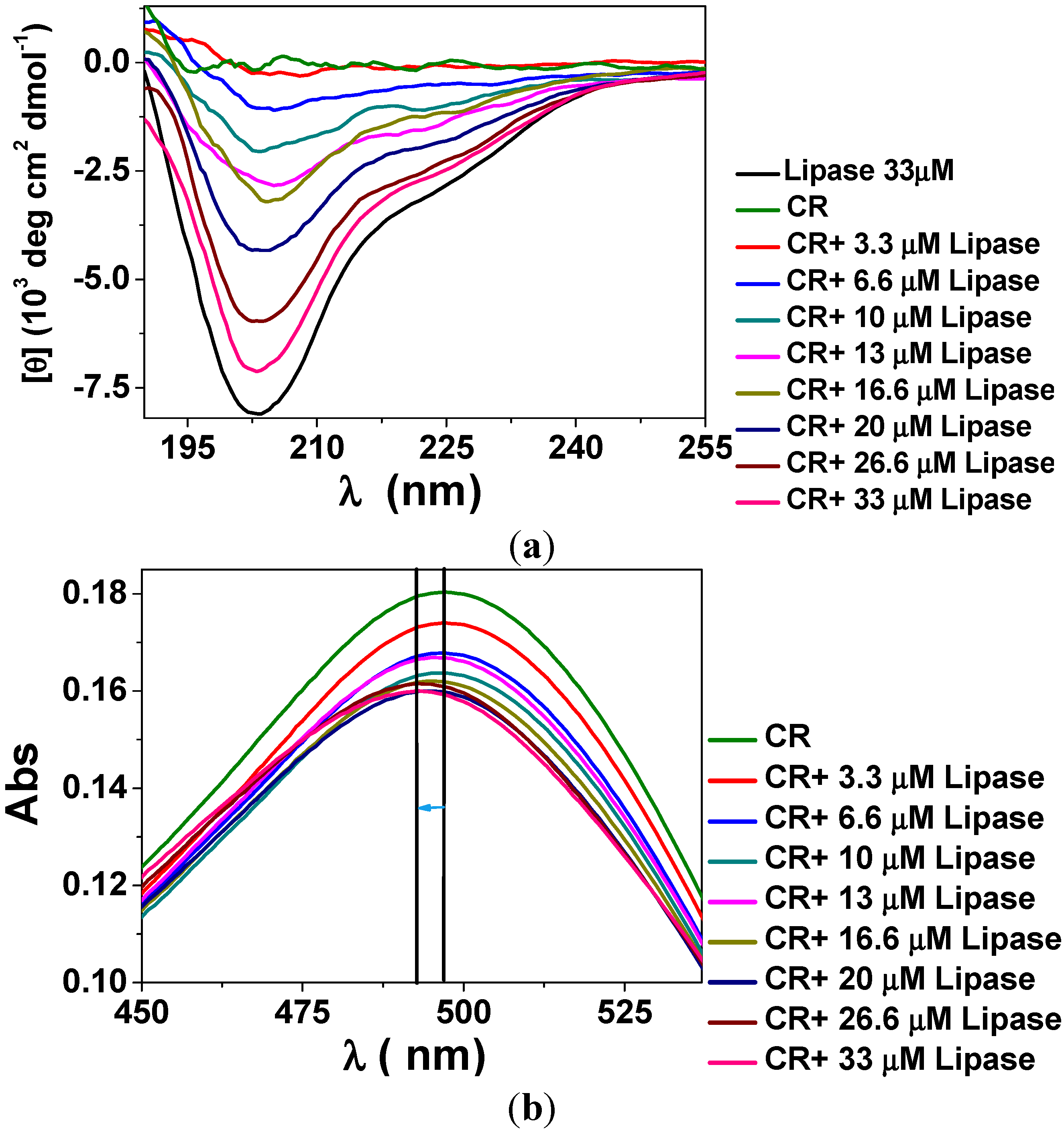

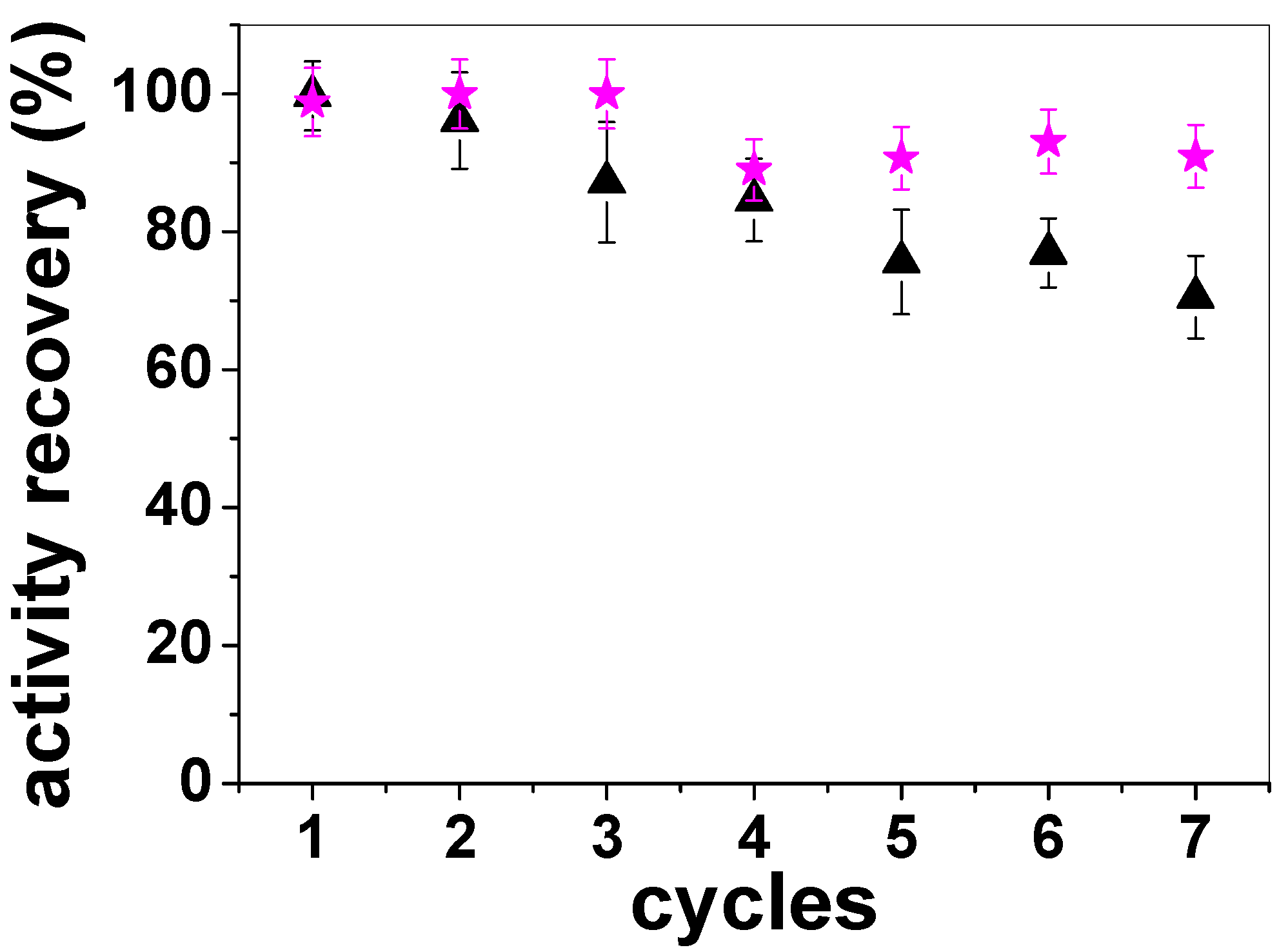
3. Experimental Section
3.1. Materials
3.2. Methods
3.2.1. Preparation of PS/PEG and PS/PEG/CR Particles
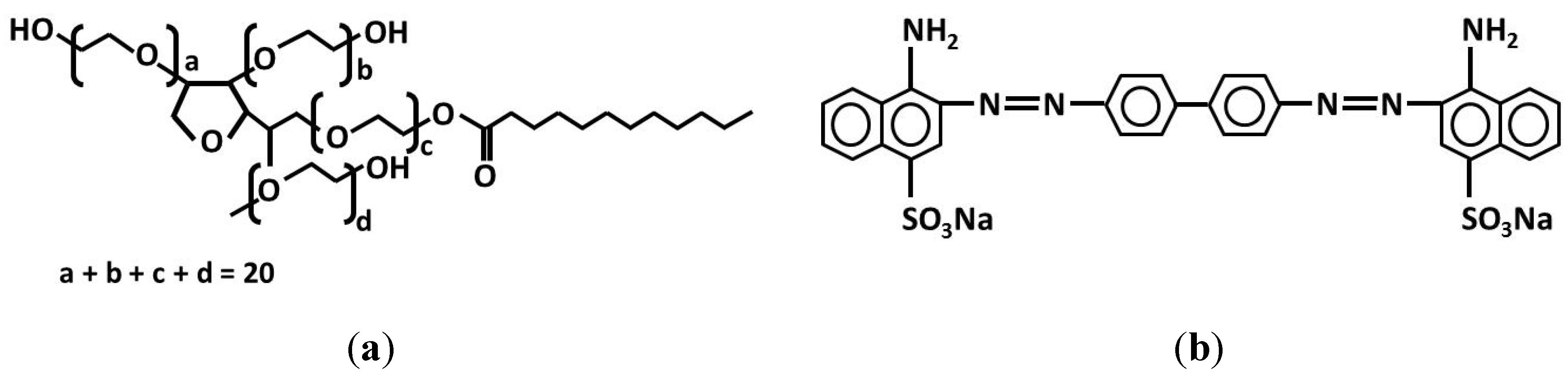
3.2.2. Adsorption Isotherms of Lipase onto PS/PEG or PS/PEG/CR Particles
3.2.3. Characterization of PS/PEG and PS/PEG/CR Particles before and after Lipase Adsorption
3.2.4. Desorption Experiments
3.2.5. Catalytic Activity of Immobilized Lipase and Free Lipase

3.2.6. Evaluation of Bioconjugation Effects
3.2.7. Ellipsometry
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Di Cosimo, R.; McAuliffe, J.; Poulose, A.J.; Bohlmann, G. Industrial use of immobilized enzymes. Chem. Soc. Rev. 2013, 42, 6437–6474. [Google Scholar]
- Kapoor, M.; Gupta, M.N. Lipase promiscuity and its biochemical applications. Proc. Biochem. 2012, 47, 555–569. [Google Scholar] [CrossRef]
- Schrag, J.D.; Cygler, M. Lipases and α/β hydrolase fold. Methods Enzymol. 1997, 284, 85–107. [Google Scholar] [CrossRef]
- Carrasco-Lopez, C.; Godoy, C.; de las Rivas, B.; Fernandez-Lorente, G.; Palomo, J.M.; Guisan, J.M. Activation of bacterial thermoalkalophilic lipases is spurred by dramatic structural rearrangements. J. Biol. Chem. 2009, 284, 4365–4372. [Google Scholar]
- Hanefeld, U.; Gardossi, L.; Magner, E. Understanding enzyme immobilisation. Chem. Soc. Rev. 2009, 38, 453–468. [Google Scholar] [CrossRef]
- Cipolatti, E.P.; Silva, M.J.A.; Klein, M.; Feddern, V.; Feltes, M.M.C.; Oliveira, J.V.; Ninow, J.L.; Oliveira, D. Current status and trends in enzymatic nanoimmobilization. J. Mol. Catal. B Enzym. 2014, 99, 56–67. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Ortiz, C.; Berenguer-Murcia, A.; Torres, R.; Fernández-Lafuente, R. Modifying enzyme activity and selectivity by immobilization. Chem. Soc. Rev. 2013, 42, 6290–6307. [Google Scholar] [CrossRef]
- Mendes, A.A.; Oliveira, P.C.; de Castro, H.F. Properties and biotechnological applications of porcine pancreatic lipase. J. Mol. Catal. BEnzym. 2012, 78, 119–134. [Google Scholar] [CrossRef]
- Sheldon, R.A.; van Pelt, S. Enzyme immobilisation in biocatalysis: Why, what and how. Chem. Soc. Rev. 2013, 42, 6223–6235. [Google Scholar] [CrossRef]
- Kosaka, P.M.; Kawano, Y.; El Seoud, O.A.; Petri, D.F.S. Catalytic activity of lipase immobilized onto ultrathin films of cellulose esters. Langmuir 2007, 23, 12167–12173. [Google Scholar] [CrossRef]
- Chen, Y.Z.; Yang, C.T.; Ching, C.B.; Xu, R. Immobilization of lipases on hydrophobilized zirconia nanoparticles: Highly enantioselective and reusable biocatalysts. Langmuir 2008, 24, 8877–8884. [Google Scholar] [CrossRef]
- Boscolo, B.; Trotta, F.; Ghibaudi, E. High catalytic performances of Pseudomonas fluorescens lipase adsorbed on a new type of cyclodextrin-based nanosponges. J. Mol. Catal. B Enzym. 2010, 62, 155–161. [Google Scholar] [CrossRef]
- Bellusci, M.; Francolini, I.; Martinelli, A.; D’Ilario, L.; Piozzi, A. Lipase immobilization on differently functionalized vinyl-based amphiphilic polymers: Influence of phase segregation on the enzyme hydrolytic activity. Biomacromolecules 2012, 13, 805–813. [Google Scholar] [CrossRef]
- Li, L.; Feng, W.; Pan, K. Immobilization of lipase on amino-cyclodextrin functionalized carbon nanotubes for enzymatic catalysis at the ionic liquid–organic solvent interface. Colloids Surf. B 2013, 102, 124–129. [Google Scholar] [CrossRef]
- Peng, Y.; Zhu-Ping, H.; Yong-Juan, X.; Peng-Cheng, H.; Ji-Jun, T. Effect of support surface chemistry on lipase adsorption and activity. J. Mol. Catal. B Enzym. 2013, 94, 69–76. [Google Scholar] [CrossRef]
- Bonfá, A.; Saito, R.S.N.; França, R.F.O.; Fonseca, B.A.L.; Petri, D.F.S. Poly(ethylene glycol) decorated poly(methylmethacrylate) nanoparticles for protein adsorption. Mater. Sci. Eng. C 2011, 31, 562–566. [Google Scholar] [CrossRef]
- Blachechen, L.S.; Silva, J.O.; Barbosa, L.R.S.; Itri, R.; Petri, D.S.F. Hofmeister effects on the colloidal stability of poly(ethylene glycol)-decorated nanoparticles. Colloid Polym. Sci. 2012, 290, 1537–1546. [Google Scholar] [CrossRef]
- Silva, R.A.; Carmona-Ribeiro, A.M.; Petri, D.F.S. Enzymatic activity of cholesterol oxidase immobilized onto polymer nanoparticles mediated by Congo red. Colloids Surf. B 2013, 110, 347–355. [Google Scholar] [CrossRef]
- Alkschbirs, M.I.; Bizotto, V.C.; Oliveira, M.G.; Sabadini, E. Effects of congo red on the drag reduction properties of Poly(ethylene oxide) in aqueous solution based on drop impact images. Langmuir 2004, 20, 11315–11320. [Google Scholar] [CrossRef]
- Cooper, T.M.; Campbell, A.L.; Crane, R.L. Formation of polypeptide-dye multilayers by an electrostatic self-assembly technique. Langmuir 1995, 11, 2713–2718. [Google Scholar] [CrossRef]
- Sharmaa, R.; Chistib, Y.; Banerjee, U.C. Production, purification, characterization, and applications of lipases. Biotechnol. Adv. 2001, 19, 627–662. [Google Scholar] [CrossRef]
- Pancera, S.M.; Itri, R.; Petri, D.F.S. Small-angle X-ray scattering on solutions of carboxymethylcellulose and bovine serum albumin. Macromol. Biosci. 2005, 5, 331–336. [Google Scholar] [CrossRef]
- Silva, R.A.; Urzúa, M.D.; Petri, D.F.S.; Dubin, P.L. Protein adsorption onto polyelectrolyte layers: Effects of protein hydrophobicity and charge anisotropy. Langmuir 2010, 26, 14032–14038. [Google Scholar] [CrossRef]
- Palomo, J.M.; Fuentes, M.; Fernández-Lorente, G.; Mateo, C.; Guisan, J.M.; Fernández-Lafuente, R. General trend of lipase to self-assemble giving bimolecular aggregates greatly modifies the enzyme functionality. Biomacromolecules 2003, 4, 1–6. [Google Scholar] [CrossRef]
- Fernández-Lorente, G.; Palomo, J.M.; Fuentes, M.; Mateo, C.; Guisán, J.M.; Fernández-Lafuente, R. Self-assembly of Pseudomonas fluorescens lipase into bimolecular aggregates dramatically affects functional properties. Biotechnol. Bioeng. 2003, 82, 232–237. [Google Scholar]
- Palomo, J.M.; Ortiz, C.; Fuentes, M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Use of immobilized lipases for lipase purification via specific lipase-lipase interactions. J. Chromatogr. A 2004, 1038, 267–273. [Google Scholar] [CrossRef]
- Palomo, J.M.; Ortiz, C.; Fernández-Lorente, G.; Fuentes, M.; Guisán, J.M.; Fernández-Lafuente, R. Lipase-lipase interactions as a new tool to immobilize and modulate the lipase properties. Enzyme Microb. Technol. 2005, 36, 447–454. [Google Scholar]
- Wilson, L.; Palomo, J.M.; Fernández-Lorente, G.; Illanes, A.; Guisán, J.M.; Fernández-Lafuente, R. Effect of lipase-lipase interactions in the activity, stability and specificity of a lipase from Alcaligenes sp. Enzyme Microb. Technol. 2006, 39, 259–264. [Google Scholar] [CrossRef]
- Azzam, R.M.A.; Bashara, N.M. Ellipsometry and Polarized Light; North Holland Personal Library: Amsterdam, The Netherlands, 1996. [Google Scholar]
- Pancera, S.M.; Gliemann, H.; Schimmel, T.; Petri, D.F.S. Effect of pH on the adsorption and activity of creatine phosphokinase. J. Phys. Chem. B 2006, 110, 2674–2680. [Google Scholar]
- Peter, G.H.; van Aalten, D.M.F.; Edfolm, O.; Toxvaerd, S.; Bywater, R. Dynamics of Proteins in Different Solvent Systems: Analysis of Essential Motion in Lipases. Biophys. J. 1996, 71, 2245–2255. [Google Scholar] [CrossRef]
- Yu, J.; Wei, X.; Zhang, L.; Fang, X.; Yang, T.; Huang, F.; Liang, W. Polyethylene glycol (PEG)-mediated conformational alteration of α-chymotrypsin prevents inactivation of insulin by stabilizing active intermediates. Mol. Pharm. 2014. [Google Scholar] [CrossRef]
- Pancera, S.M.; Petri, D.F.S.; Itri, R. The effect of poly(ethylene glycol) on the activity, structural conformation and stability of yeast hexokinase. Prog. Colloid Polym. Sci. 2004, 128, 178–183. [Google Scholar]
- Pancera, S.M.; da Silva, L.H.M.; Loh, W.; Itri, R.; Pessoa, A., Jr.; Petri, D.F.S. The effect of Poly(ethylene glycol) on the activity and structure of glucose-6-phosphate dehydrogenase in solution. Colloids Surf. B 2002, 26, 291–300. [Google Scholar] [CrossRef]
- Polizelli, P.P.; Tiera, M.J.; Bonilla-Rodriguez, G.O. Effect of surfactants and polyethylene glycol on the activity and stability of a lipase from oilseeds of Pachira aquatica. J. Am. Oil Chem. Soc. 2008, 85, 749–753. [Google Scholar] [CrossRef]
- Bayramoglu, G.; Hazer, B.; Altıntas, B.; Arıca, M.Y. Covalent immobilization of lipase onto amine functionalized polypropylene membrane and its application in green apple flavor (ethyl valerate) synthesis. Proc. Biochem. 2011, 46, 372–378. [Google Scholar]
- Hatzinikolaou, D.G.; Kourentzi, E.; Stamatis, H.; Christakopoulos, P.; Kolisis, F.N.; Kekos, D.; Macris, B.J. A novel lipolytic activity of Rhodotorula glutinis cells: Production, partial characterization and application in the synthesis of esters. J. Biosci. Bioeng. 1999, 88, 53–56. [Google Scholar]
- Mateo, C.; Palono, J.M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Improvementof enzyme activity, stability and selectivity via immobilization techniques. Enzym. Microb. Technol. 2007, 40, 1451–1463. [Google Scholar] [CrossRef]
- Dodson, G.G.; Lawson, D.M.; Winkler, F.K. Structural and evolutionary relationships in lipase mechanism and activation. Faraday Discuss. 1992, 93, 95–105. [Google Scholar] [CrossRef]
- Fabian, J.; Hartmann, H. Light Absorption of Organic Colorants; Springer-Verlag: Berlin, Germany, 1980. [Google Scholar]
- Pereira, E.B.; Zanin, G.M.; Castro, H.F. Immobilization and catalytic properties of lipase on chitosan for hydrolysis and esterification reactions. J. Braz. Chem. Eng. 2003, 20, 343–355. [Google Scholar]
- Benjamin, S.; Pandey, A. Isolation and characterization of three distinct forms of lipases from Candida rugosa produced in solid state fermentation. Braz. Arch. Biol. Technol. 2000, 43, 453–460. [Google Scholar] [CrossRef]
- Grabowski, E.F.; Morrison, I.D. Particle size distribution from analysis of quasielastic light scattering data. In Measurements of Suspended Particles by Quasi-Elastic Light Scattering; Dahneke, B.E., Ed.; Wiley: New York, NY, USA, 1983; pp. 199–236. [Google Scholar]
- Castro, L.B.R.; Soares, K.V.; Naves, A.F.; Carmona-Ribeiro, A.M.; Petri, D.F.S. Synthesis of stable polystyrene and poly(methyl methacrylate) particles in the presence of carboxymethyl Cellulose. Ind. Eng. Chem. Res. 2004, 43, 7774–7779. [Google Scholar] [CrossRef]
- Iacazio, G.; Périssol, C.; Faure, B. A new tannase substrate for spectrophotometric assay. J. Microbiol. Methods 2000, 42, 209–214. [Google Scholar] [CrossRef]
- Sample Availability: Colloidal dispersions of PS/PEG, PS/PEG/CR and PS/PEG/CR coated with lipases are available from the authors.
© 2014 by the authors. licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Silva, R.A.; Carmona-Ribeiro, A.M.; Petri, D.F.S. Catalytic Behavior of Lipase Immobilized onto Congo Red and PEG-Decorated Particles. Molecules 2014, 19, 8610-8628. https://doi.org/10.3390/molecules19068610
Silva RA, Carmona-Ribeiro AM, Petri DFS. Catalytic Behavior of Lipase Immobilized onto Congo Red and PEG-Decorated Particles. Molecules. 2014; 19(6):8610-8628. https://doi.org/10.3390/molecules19068610
Chicago/Turabian StyleSilva, Rubens A., Ana M. Carmona-Ribeiro, and Denise F. S. Petri. 2014. "Catalytic Behavior of Lipase Immobilized onto Congo Red and PEG-Decorated Particles" Molecules 19, no. 6: 8610-8628. https://doi.org/10.3390/molecules19068610





