Cloning and Characterisation of the Gene Encoding 3-Hydroxy-3-Methylglutaryl-CoA Synthase in Tripterygium wilfordii
Abstract
:1. Introduction
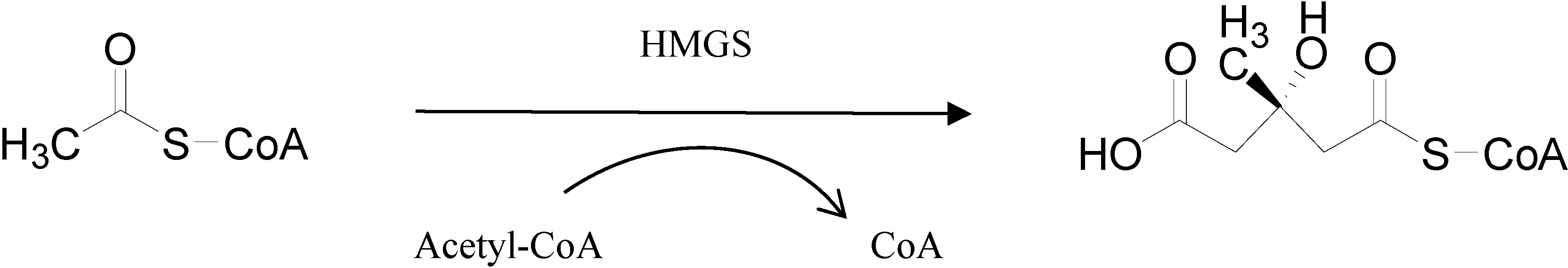
2. Results and Discussion
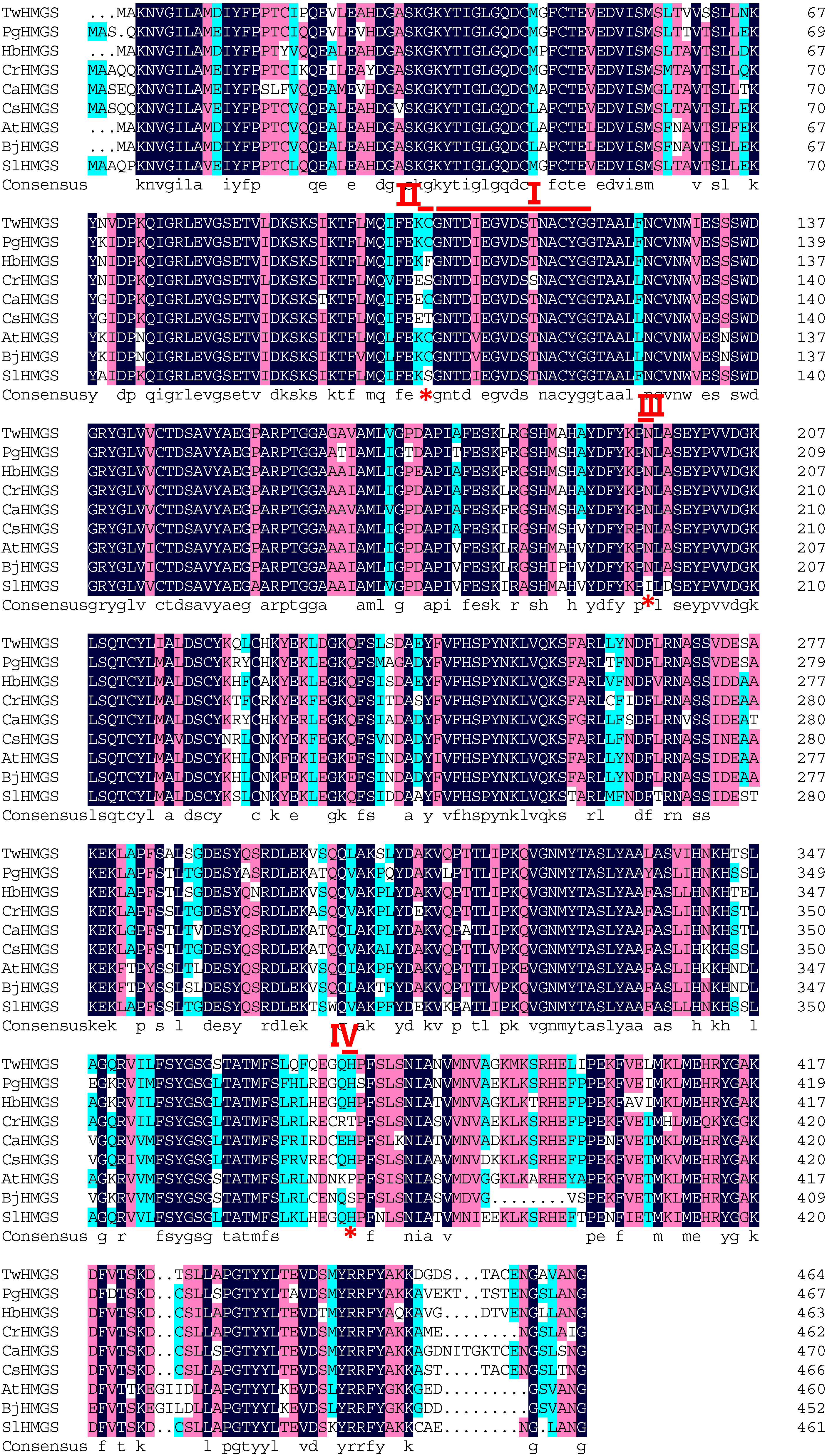
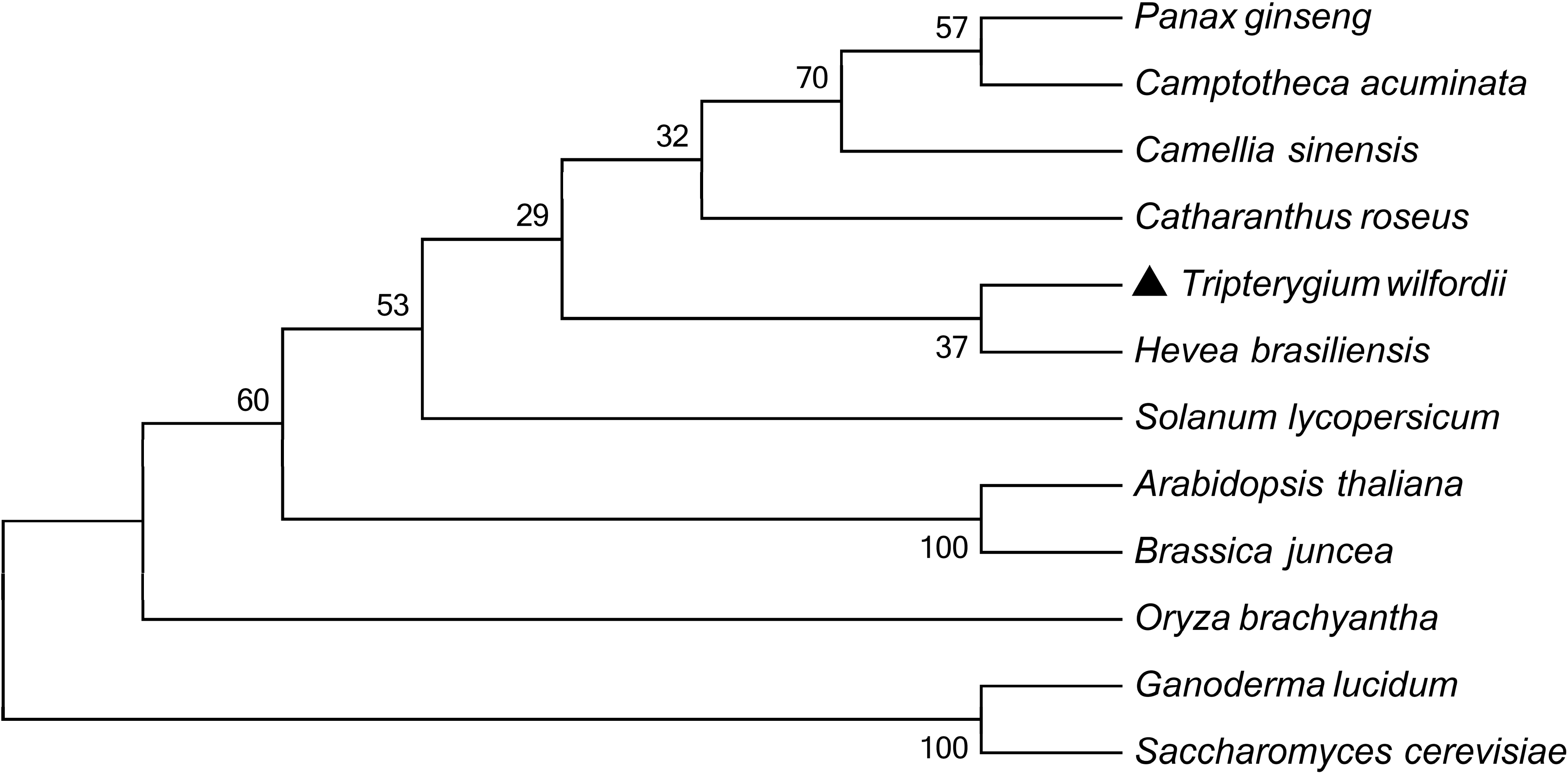
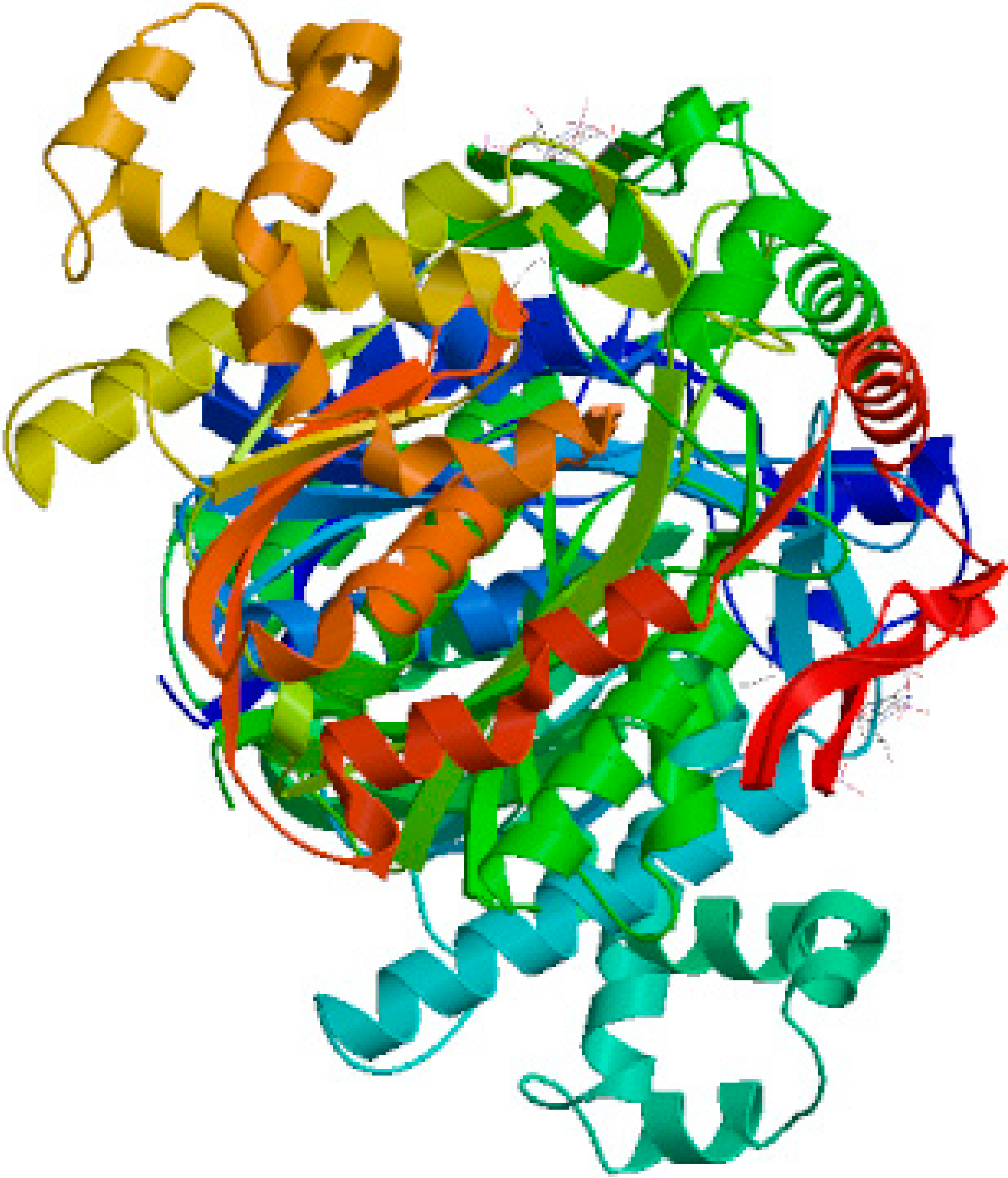
2.1. Cloning and Sequence Analysis of TwHMGS
2.2. Functional Identification in Yeast

2.3. Expression of TwHMGS in the Suspension Cells

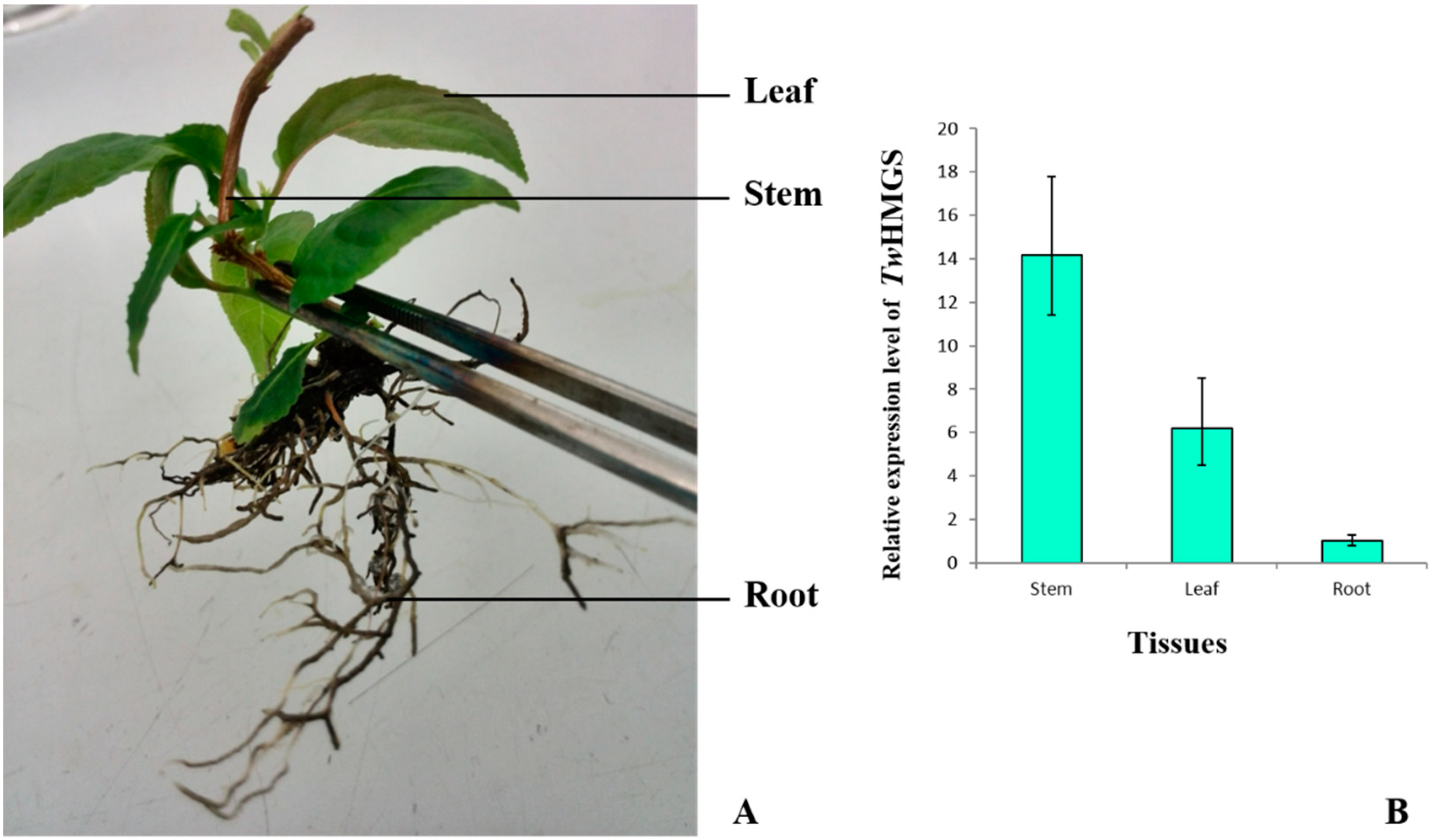
2.4. Expression of TwHMGS in Tissues
2.5. Discussion
3. Experimental Section
3.1. Plant Material
3.2. RNA Isolation
3.3. Strains
3.4. Cloning of TwHMGS Full-Length cDNA
3.5. Bioinformatics Analysis
3.6. Construction of the pYES2-TwHMGS Expression Vector
3.7. Complementation Assays
3.8. Quantitative Real-Time PCR
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wang, C.; Li, C.J.; Yang, J.Z.; Ma, J.; Chen, X.G.; Hou, Q.; Zhang, D.M. Anti-inflammatory sesquiterpene derivatives from the leaves of Tripterygium. wilfordii. J. Nat. Prod. 2013, 76, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Marcus, D.M. Comparison of Tripterygium wilfordii Hook F with methotrexate in the treatment of rheumatoid arthritis. Ann. Rheum. Dis. 2014, 73. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Huang, W.; Ousman, T.; Zhang, B.; Han, Y.; Clotaire, D.Z.; Wang, C.; Chang, H.; Luo, H.; Ren, X.; et al. Triptolide induces growth inhibition and apoptosis of human laryngocarcinoma cells by enhancing p53 activities and suppressing E6-mediated p53 degradation. PLoS One 2013, 8, e80784. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Zhang, X.; Zhao, M.; Wang, Y.; Cheng, X.; Wang, D.; Xu, Y.; Du, Z.; Yu, X. Celastrol targets mitochondrial respiratory chain complex I to induce reactive oxygen species-dependent cytotoxicity in tumor cells. BMC Cancer 2011, 11. [Google Scholar] [CrossRef]
- Su, P.; Cheng, Q.; Wang, X.; Cheng, X.; Zhang, M.; Tong, Y.; Li, F.; Gao, W.; Huang, L. Characterization of eight terpenoids from tissue cultures of the Chinese herbal plant, Tripterygium wilfordii, by high-performance liquid chromatography coupled with electrospray ionization tandem mass spectrometry. Biomed. Chromatogr. 2014, 28, 1183–1192. [Google Scholar] [CrossRef] [PubMed]
- Yan, X; Ke, X.X.; Zhao, H.; Huang, M.; Hu, R.; Cui, H. Triptolide inhibits cell proliferation and tumorigenicity of human neuroblastoma cells. Mol. Med. Rep. 2014. [Google Scholar] [CrossRef]
- Han, X.; Sun, S.; Zhao, M.; Cheng, X.; Chen, G.; Lin, S.; Guan, Y.; Yu, X. Celastrol stimulates hypoxia-inducible factor-1 activity in tumor cells by initiating the ROS/Akt/p70S6K signaling pathway and enhancing hypoxia-inducible factor-1α protein synthesis. PLoS One 2014, 9, e112470. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.G.; Song, L.; Liu, M.; Hu, Z.B.; Wang, Z.T. Advancement in analysis of Salviae miltiorrhizae Radix et Rhizoma (Danshen). J. Chromatogr. A 2009, 1216, 1941–1953. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Ding, C.; Ge, Q.; Zhou, Z.; Zhi, X. Simultaneous determination of ginkgolides A, B, C and bilobalide in plasma by LC-MS/MS and its application to the pharmacokinetic study of Ginkgo biloba extract in rats. J. Chromatogr. B 2008, 864, 87–94. [Google Scholar] [CrossRef]
- Lu, L.; Zhao, Y.; Yu, H.S.; Huang, H.Z.; Kang, L.P.; Cao, M.; Cui, J.M.; Yu, L.Y.; Song, X.B.; Ma, B.P. Preparation of Glycyrrhetinic Acid Monoglucuronide by Selective Hydrolysis of Glycyrrhizic Acid via Biotransformation. Chin. Herbal Med. 2012, 4, 324–328. [Google Scholar]
- Laule, O.; Fürholz, A.; Chang, H.S.; Zhu, T.; Wang, X.; Heifetz, P.B.; Gruissem, W.; Lange, M. Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc. Nat. Acad. Sci. USA 2003, 100, 6866–6871. [Google Scholar] [CrossRef] [PubMed]
- Chappell, J. Biochemistry and molecular biology of the isoprenoid biosynthetic pathway in plants. Plant Mol. Biol. 1995, 46, 521–547. [Google Scholar]
- Denbow, C.J.; Lång, S.; Cramer, C.L. The N-terminal domain of tomato 3-hydroxy-3-methylglutaryl-CoA reductases. Sequence, microsomal targeting, and glycosylation. J. Biol. Chem. 1996, 271, 9710–9715. [Google Scholar]
- Skorupinska-Tudek, K.; Poznanski, J.; Wojcik, J.; Bienkowski, T.; Szostkiewicz, I.; Zelman-Femiak, M.; Bajda, A.; Chojnacki, T.; Olszowska, O.; Grunler, J.; et al. Contribution of the mevalonate and methylerythritol phosphate pathways to the biosynthesis of dolichols in plants. J. Biol. Chem. 2008, 283, 21024–21035. [Google Scholar] [CrossRef] [PubMed]
- Opitz, S.; Nes, W.D.; Gershenzon, J. Both methylerythritol phosphate and mevalonate pathways contribute to biosynthesis of each of the major isoprenoid classes in young cotton seedlings. Phytochemistry 2014, 98, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Chun, K.Y.; Vinarov, D.A.; Zajicek, J.; Miziorko, H.M. 3-Hydroxy-3-methylglutaryl-CoA synthase. A role for glutamate 95 in general acid/base catalysis of C-C bond formation. J. Biol. Chem. 2000, 275, 17946–17953. [Google Scholar] [CrossRef] [PubMed]
- Greenspan, M.D.; Yudkovitz, J.B.; Lo, C.Y.; Chen, J.S.; Alberts, A.W.; Hunt, V.M.; Chang, M.N.; Yang, S.S.; Thompson, K.L.; Chiang, Y.C.; et al. Inhibition of hydroxymethylglutaryl-coenzyme A synthase by L-659,699. Proc. Nat. Acad. Sci. USA 1987, 84, 7488–7492. [Google Scholar] [CrossRef]
- Theisen, M.J.; Misra, I.; Saadat, D.; Campobasso, N.; Miziorko, H.M.; Harrison, D.H. 3-Hydroxy-3-methylglutaryl-CoA synthase intermediate complex observed in “real-time”. Proc. Nat. Acad. Sci. USA 2004, 101, 16442–16447. [Google Scholar] [CrossRef] [PubMed]
- Vallett, S.M.; Sanchez, H.B.; Rosenfeld, J.M.; Osborne, T.F. A direct role for sterol regulatory element binding protein in activation of 3-hydroxy-3-methylglutaryl coenzyme A reductase gene. J. Biol. Chem. 1996, 271, 12247–12253. [Google Scholar] [CrossRef] [PubMed]
- Dooley, K.A.; Millinder, S.; Osborne, T.F. Sterol regulation of 3-hydroxy-3-methylglutaryl-coenzyme A synthase gene through a direct interaction between sterol regulatory element binding protein and the trimeric CCAAT-binding factor/nuclear factor Y. J. Biol. Chem. 1998, 273, 1349–1356. [Google Scholar] [CrossRef] [PubMed]
- Pojer, F.; Ferrer, J.L.; Richard, S.B.; Nagegowda, D.A.; Chye, M.L.; Bach, T.J.; Noel, J.P. Structural basis for the design of potent and species-specific inhibitors of 3-hydroxy-3-methylglutaryl CoA synthases. Proc. Nat. Acad. Sci. USA 2006, 103, 11491–11496. [Google Scholar] [CrossRef] [PubMed]
- Sando, T.; Takeno, S.; Watanabe, N.; Okumoto, H.; Kuzuyama, T.; Yamashita, A.; Hattori, M.; Ogasawara, N.; Fukusaki, E.; Kobayashi, A. Cloning and characterization of mevalonate pathway genes in a natural rubber producing plant, Hevea brasiliensis. Biosci. Biotechnol. Biochem. 2008, 72, 2049–2060. [Google Scholar] [CrossRef] [PubMed]
- Kai, G.; Miao, Z.; Zhang, L.; Zhao, D.; Liao, Z.; Sun, X.; Zhao, L.; Tang, K. Molecular cloning and expression analyses of a new gene encoding 3-hydroxy-3-methylglutaryl-CoA synthase from Taxus media. Biol. Plant. 2006, 50, 359–366. [Google Scholar] [CrossRef]
- Montamat, F.; Guilloton, M.; Karst, F.; Delrot, S. Isolation and characterization of a cDNA encoding Arabidopsis thaliana 3-hydroxy-3-methylglutaryl-coenzyme A synthase. Gene 1995, 167, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Price, A.C.; Zhang, Y.M.; Rock, C.O.; White, S.W. Structure of beta-ketoacyl-[acyl carrier protein] reductase from Escherichia coli: negative cooperativity and its structural basis. Biochemistry 2001, 40, 12772–12781. [Google Scholar] [CrossRef] [PubMed]
- Alper, H.; Miyaoku, K.; Stephanopoulos, G. Construction of lycopene-overproducing E.coli strains by combining systematic and combinatorial gene knockout targets. Nat. Biotechol. 2005, 23, 612–616. [Google Scholar] [CrossRef]
- Ajikumar, P.K.; Xiao, W.H.; Tyo, K.E.; Wang, Y.; Simeon, F.; Leonard, E.; Mucha, O.; Phon, T.H.; Pfeifer, B.; Stephanopoulos, G. Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli. Science 2010, 220, 70–74. [Google Scholar] [CrossRef] [Green Version]
- Del Sal, G.; Manfioletti, G.; Schneider, C. The CTAB-DNA precipitation method: A common mini-scale preparation of template DNA from phagemids, phages or plasmids suitable for sequencing. Biotechniques 1989, 7, 514–520. [Google Scholar] [PubMed]
- ORF Finder (Open Reading Frame Finder). Available online: http://www.ncbi.nlm.nih.gov/projects/gorf/ (accessed on 25 November 2014).
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using realtime quantitative PCR and the 2-ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds are available from the authors.
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.-J.; Zhao, Y.-J.; Zhang, M.; Su, P.; Wang, X.-J.; Zhang, X.-N.; Gao, W.; Huang, L.-Q. Cloning and Characterisation of the Gene Encoding 3-Hydroxy-3-Methylglutaryl-CoA Synthase in Tripterygium wilfordii. Molecules 2014, 19, 19696-19707. https://doi.org/10.3390/molecules191219696
Liu Y-J, Zhao Y-J, Zhang M, Su P, Wang X-J, Zhang X-N, Gao W, Huang L-Q. Cloning and Characterisation of the Gene Encoding 3-Hydroxy-3-Methylglutaryl-CoA Synthase in Tripterygium wilfordii. Molecules. 2014; 19(12):19696-19707. https://doi.org/10.3390/molecules191219696
Chicago/Turabian StyleLiu, Yu-Jia, Yu-Jun Zhao, Meng Zhang, Ping Su, Xiu-Juan Wang, Xia-Nan Zhang, Wei Gao, and Lu-Qi Huang. 2014. "Cloning and Characterisation of the Gene Encoding 3-Hydroxy-3-Methylglutaryl-CoA Synthase in Tripterygium wilfordii" Molecules 19, no. 12: 19696-19707. https://doi.org/10.3390/molecules191219696



