Influence of Disulfide Connectivity on Structure and Bioactivity of α-Conotoxin TxIA
Abstract
:1. Introduction

2. Results and Discussion
2.1. Results
2.1.1. Purification and Identification of α-CTx TxIA from C. textile
2.1.2. Synthesis of α-CTx TxIA Isomers by Two-Step Oxidation
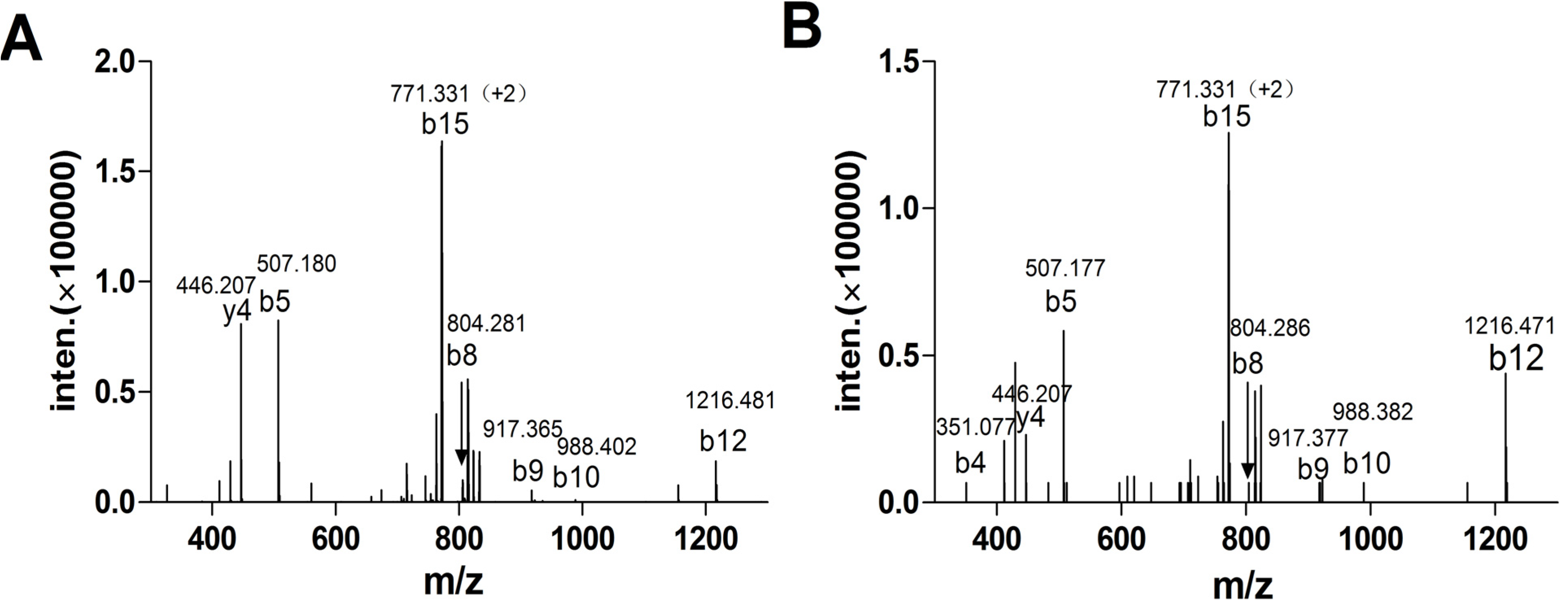
2.1.3. Peptide Synthesis of α-CTx TxIA by One-Step Oxidation

2.1.4. α-CTx TxIA Isomers Exhibit Differential Activity Against α3β2 nAChRs

2.1.5. Circular Dichroism (CD) Analysis of α-CTx TxIA Isomers

2.2. Discussion
3. Experimental
3.1. Materials
3.2. Venom Fractionation
3.3. Tandem Mass Spectrometric Analysis
3.4. Chemical Synthesis of α-CTx TxIA by Two-Step Oxidation
3.5. Chemical Synthesis of α-CTx TxIA by One-Step Oxidation
3.6. Reduction and Reoxidation of Native α-CTx TxIA
3.7. RNA Preparation
3.8. Voltage Clamp Recording and Data Analysis
3.9. Circular Dichroism (CD) Spectroscopy
4. Conclusions
Abbreviations
| AChBPs | Acetylcholine binding proteins |
| Acm | Acetamidomethyl |
| ACN | acetonitrile |
| CTx | conotoxin |
| CD | Circular dichroism |
| EDTA | ethylenediaminetetraacetic acid |
| ESI-MS | electrospray ionization-mass spectrometry |
| Fmoc | 9-fluorenylmethyloxycarbonyl |
| IT-TOF MS | ion trap time-of-flight mass spectrometer |
| nAChRs | nicotinic acetylcholine receptors |
| OtBu | tertiary butyl ester |
| Pbf | 2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl |
| RP-HPLC | Reversed-phase high-performance liquid chromatography |
| tBu | t-Butyl |
| TFA | trifluoroacetic acid |
| Trt | Trityl |
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Terlau, H.; Olivera, B.M. Conus venoms: A rich source of novel ion channel-targeted peptides. Physiol. Rev. 2004, 84, 41–68. [Google Scholar] [CrossRef]
- Armishaw, C.J.; Alewood, P.F. Conotoxins as research tools and drug leads. Curr. Protein Pept. Sci. 2005, 6, 221–240. [Google Scholar] [CrossRef]
- Lewis, R.J.; Dutertre, S.; Vetter, I.; Christie, M.J. Conus venom Peptide pharmacology. Physiol. Rev. 2012, 64, 259–298. [Google Scholar]
- Millard, E.L.; Daly, N.L.; Craik, D.J. Structure-activity relationships of alpha-conotoxins targeting neuronal nicotinic acetylcholine receptors. FEBS J. 2004, 271, 2320–2326. [Google Scholar]
- Essack, M.; Bajic, V.B.; Archer, J.A. Conotoxins that confer therapeutic possibilities. Mar. Drugs 2012, 10, 1244–1265. [Google Scholar] [CrossRef]
- Gotti, C.; Moretti, M.; Bohr, I.; Ziabreva, I.; Vailati, S.; Longhi, R.; Riganti, L.; Gaimarri, A.; McKeith, I.G.; Perry, R.H.; et al. Selective nicotinic acetylcholine receptor subunit deficits identified in Alzheimer’s disease, Parkinson’s disease and dementia with Lewy bodies by immunoprecipitation. Neurobiol. Dis. 2006, 23, 481–489. [Google Scholar] [CrossRef]
- Gotti, C.; Clementi, F. Neuronal nicotinic receptors: From structure to pathology. Prog. Neurobiol. 2004, 74, 363–396. [Google Scholar] [CrossRef]
- Dutertre, S.; Ulens, C.; Buttner, R.; Fish, A.; van Elk, R.; Kendel, Y.; Hopping, G.; Alewood, P.F.; Schroeder, C.; Nicke, A.; et al. AChBP-targeted alpha-conotoxin correlates distinct binding orientations with nAChR subtype selectivity. EMBO J. 2007, 26, 3858–3867. [Google Scholar] [CrossRef]
- Dobson, R.; Collodoro, M.; Gilles, N.; Turtoi, A.; de Pauw, E.; Quinton, L. Secretion and maturation of conotoxins in the venom ducts of Conus textile. Toxicon 2012, 60, 1370–1379. [Google Scholar] [CrossRef]
- Gyanda, R.; Banerjee, J.; Chang, Y.P.; Phillips, A.M.; Toll, L.; Armishaw, C.J. Oxidative folding and preparation of alpha-conotoxins for use in high-throughput structure-activity relationship studies. J. Pept. Sci. 2013, 19, 16–24. [Google Scholar] [CrossRef]
- Jin, A.H.; Brandstaetter, H.; Nevin, S.T.; Tan, C.C.; Clark, R.J.; Adams, D.J.; Alewood, P.F.; Craik, D.J.; Daly, N.L. Structure of alpha-conotoxin BuIA: Influences of disulfide connectivity on structural dynamics. BMC Struct. Biol. 2007, 7, 28. [Google Scholar] [CrossRef]
- Gehrmann, J.; Alewood, P.F.; Craik, D.J. Structure determination of the three disulfide bond isomers of alpha-conotoxin GI: A model for the role of disulfide bonds in structural stability. J. Mol. Biol. 1998, 278, 401–415. [Google Scholar] [CrossRef]
- Zhang, R.M.; Snyder, G.H. Factors governing selective formation of specific disulfides in synthetic variants of alpha-conotoxin. Biochemistry 1991, 30, 11343–11348. [Google Scholar] [CrossRef]
- Nielsen, J.S.; Buczek, P.; Bulaj, G. Cosolvent-assisted oxidative folding of a bicyclic alpha-conotoxin ImI. J. Pept. Sci. 2004, 10, 249–256. [Google Scholar] [CrossRef]
- Davis, J.; Jones, A.; Lewis, R.J. Remarkable inter- and intra-species complexity of conotoxins revealed by LC/MS. Peptides 2009, 30, 1222–1227. [Google Scholar]
- Tayo, L.L.; Lu, B.; Cruz, L.J.; Yates, J.R., 3rd. Proteomic analysis provides insights on venom processing in Conus textile. J. Proteome Res. 2010, 9, 2292–2301. [Google Scholar] [CrossRef]
- Kaas, Q.; Westermann, J.C.; Halai, R.; Wang, C.K.; Craik, D.J. ConoServer, a database for conopeptide sequences and structures. Bioinformatics 2008, 24, 445–446. [Google Scholar] [CrossRef]
- Kaas, Q.; Yu, R.; Jin, A.H.; Dutertre, S.; Craik, D.J. ConoServer: Updated content, knowledge, and discovery tools in the conopeptide database. Nucleic Acids Res. 2012, 40, D325–D330. [Google Scholar] [CrossRef]
- Dani, J.A.; Bertrand, D. Nicotinic acetylcholine receptors and nicotinic cholinergic mechanisms of the central nervous system. Ann. Rev. Pharmacol. Toxicol. 2007, 47, 699–729. [Google Scholar] [CrossRef]
- Wallace, T.L.; Bertrand, D. Alpha7 neuronal nicotinic receptors as a drug target in schizophrenia. Exp. Opin. Therap. Tar. 2013, 17, 139–155. [Google Scholar] [CrossRef]
- Dutton, J.L.; Bansal, P.S.; Hogg, R.C.; Adams, D.J.; Alewood, P.F.; Craik, D.J. A new level of conotoxin diversity, a non-native disulfide bond connectivity in alpha-conotoxin AuIB reduces structural definition but increases biological activity. J. Biol. Chem. 2002, 277, 48849–48857. [Google Scholar]
- Balaji, R.A.; Ohtake, A.; Sato, K.; Gopalakrishnakone, P.; Kini, R.M.; Seow, K.T.; Bay, B.H. Lambda-conotoxins, a new family of conotoxins with unique disulfide pattern and protein folding. Isolation and characterization from the venom of Conus marmoreus. J. Biol. Chem. 2000, 275, 39516–39522. [Google Scholar]
- Sharpe, I.A.; Gehrmann, J.; Loughnan, M.L.; Thomas, L.; Adams, D.A.; Atkins, A.; Palant, E.; Craik, D.J.; Adams, D.J.; Alewood, P.F.; et al. Two new classes of conopeptides inhibit the alpha1-adrenoceptor and noradrenaline transporter. Nat. Neurosci. 2001, 4, 902–907. [Google Scholar] [CrossRef]
- Lovelace, E.S.; Gunasekera, S.; Alvarmo, C.; Clark, R.J.; Nevin, S.T.; Grishin, A.A.; Adams, D.J.; Craik, D.J.; Daly, N.L. Stabilization of alpha-conotoxin AuIB: Influences of disulfide connectivity and backbone cyclization. Antioxid. Redox. Sign. 2011, 14, 87–95. [Google Scholar] [CrossRef]
- Luo, S.; Akondi, K.B.; Zhangsun, D.; Wu, Y.; Zhu, X.; Hu, Y.; Christensen, S.; Dowell, C.; Daly, N.L.; Craik, D.J.; et al. Atypical alpha-conotoxin LtIA from Conus litteratus targets a novel microsite of the alpha3beta2 nicotinic receptor. J. Biol. Chem. 2010, 285, 12355–12366. [Google Scholar] [CrossRef]
- Sample Availability: Samples of the compounds are available from the authors.
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wu, Y.; Wu, X.; Yu, J.; Zhu, X.; Zhangsun, D.; Luo, S. Influence of Disulfide Connectivity on Structure and Bioactivity of α-Conotoxin TxIA. Molecules 2014, 19, 966-979. https://doi.org/10.3390/molecules19010966
Wu Y, Wu X, Yu J, Zhu X, Zhangsun D, Luo S. Influence of Disulfide Connectivity on Structure and Bioactivity of α-Conotoxin TxIA. Molecules. 2014; 19(1):966-979. https://doi.org/10.3390/molecules19010966
Chicago/Turabian StyleWu, Yong, Xiaosa Wu, Jinpeng Yu, Xiaopeng Zhu, Dongting Zhangsun, and Sulan Luo. 2014. "Influence of Disulfide Connectivity on Structure and Bioactivity of α-Conotoxin TxIA" Molecules 19, no. 1: 966-979. https://doi.org/10.3390/molecules19010966




