Inhibition of Epstein-Barr Virus Lytic Cycle by an Ethyl Acetate Subfraction Separated from Polygonum cuspidatum Root and Its Major Component, Emodin
Abstract
:1. Introduction
2. Results and Discussion
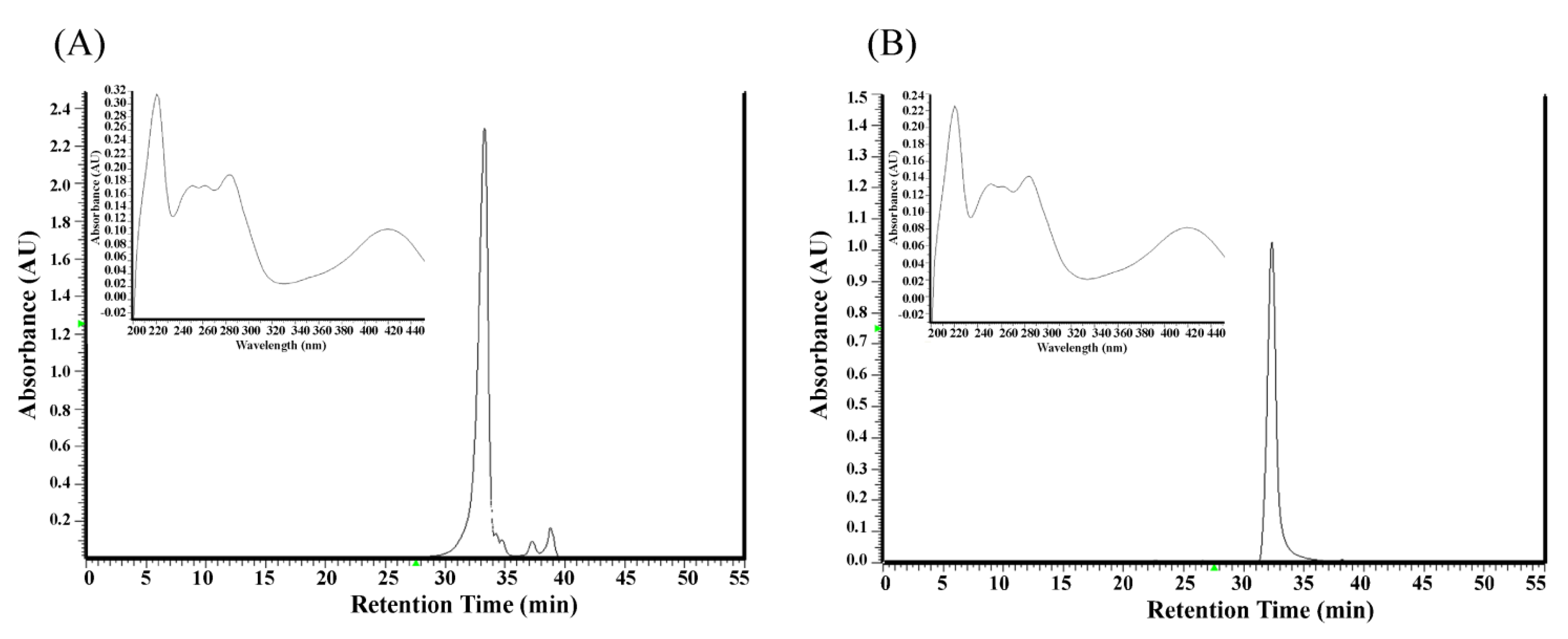
2.1. Analysis of Components in the Ethyl Acetate Subfraction F3 Isolated from Polygonum cuspidatum
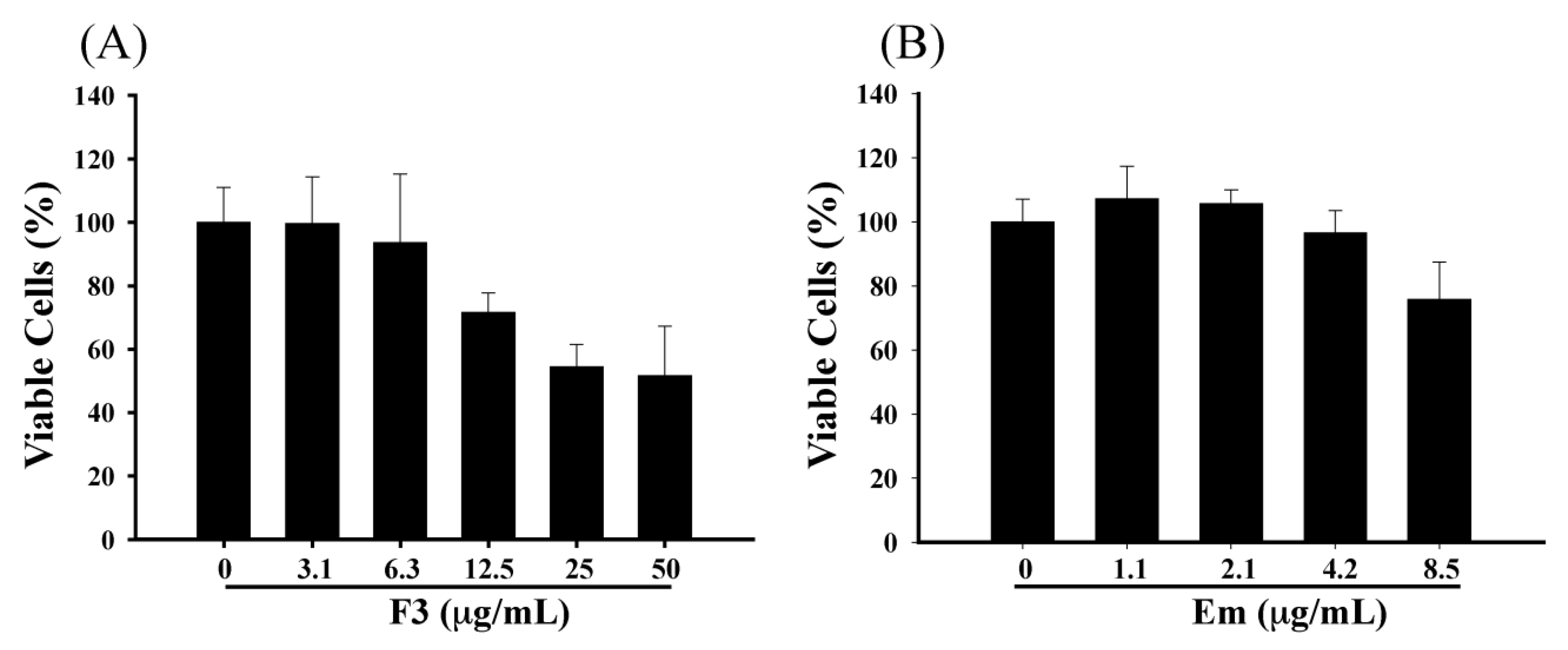
2.2. Effect of F3 and Emodin on the Viability of P3HR1 Cells
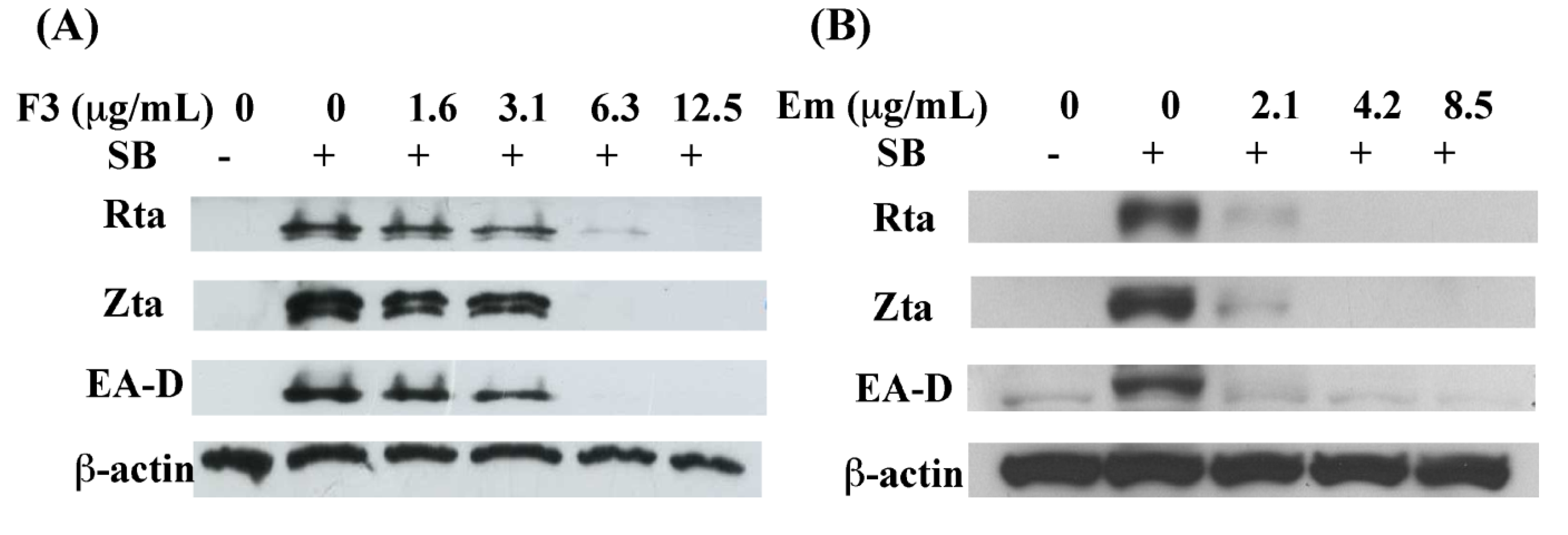
2.3. Inhibition of EBV Lytic Proteins Expression by F3
2.4. Indirect Immunofluorescence Analysis of EBV Lytic Proteins Expression

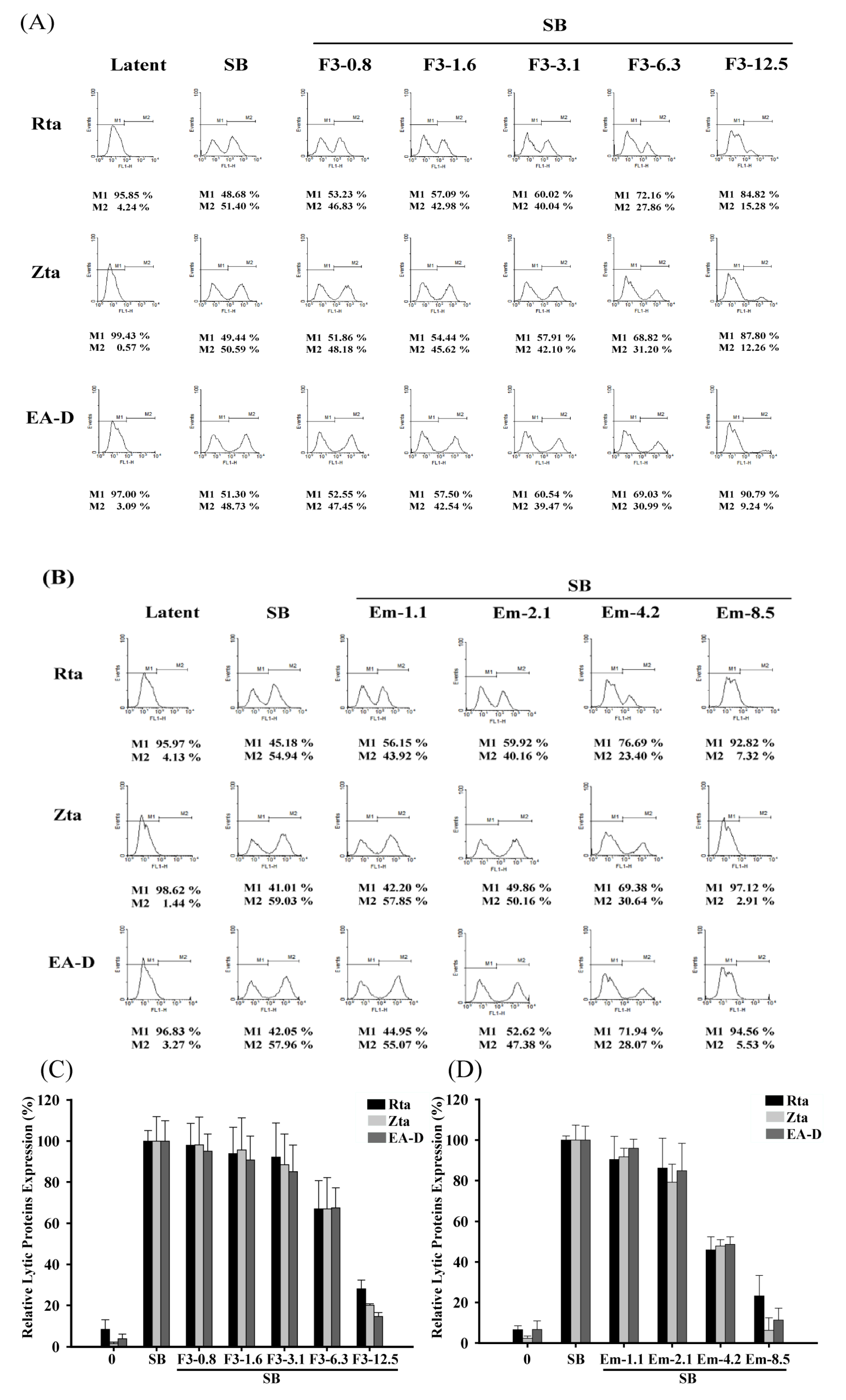
2.5. Quantitative Flow Cytometry Analysis of the Inhibitory Effects of F3 on EBV Lytic Proteins Expression
2.6. Inhibiting the Transcription of EBV Immediate-Early Genes

2.7. Inhibition of the EBV DNA Replication

2.8. Discussion
3. Experimental
3.1. Material
3.2. Sample Preparation
3.3. HPLC Analysis
3.4. Cell Culture and Lytic Induction of EBV
3.5. Cell Viability Assay
3.6. Immunoblot Analysis
3.7. Indirect Immunofluorescence Staining
3.8. Flow Cytometric Analysis
3.9. RNA and DNA Extraction
3.10. Real-Time Quantitative PCR
3.11. Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Roizman, B.; Carmichael, L.E.; Deinhardt, F.; de-The, G.; Nahmias, A.J.; Plowright, W.; Rapp, F.; Sheldrick, P.; Takahashi, M.; Wolf, K. Herpesviridae. Definition, provisional nomenclature, and taxonomy. Intervirology 1981, 16, 201–217. [Google Scholar] [CrossRef]
- Magrath, I.; Jain, V.; Bhatia, K. Epstein-Barr virus and Burkitt's lymphoma. Semin. Cancer Biol. 1992, 3, 285–295. [Google Scholar]
- Zur Hausen, H.; Schulte-Holthausen, H.; Klein, G.; Henle, W.; Henle, G.; Clifford, P.; Santesson, L. EBV DNA in biopsies of Burkitt tumours and anaplastic carcinomas of the nasopharynx. Nature 1970, 228, 1056–1058. [Google Scholar] [CrossRef]
- Weiss, L.M.; Movahed, L.A.; Warnke, R.A.; Sklar, J. Detection of Epstein-Barr viral genomes in Reed-Sternberg cells of Hodgkin’s disease. N. Engl. J. Med. 1989, 320, 502–506. [Google Scholar] [CrossRef]
- Shibata, D.; Weiss, L.M. Epstein-Barr virus-associated gastric adenocarcinoma. Am. J. Pathol. 1992, 140, 769–774. [Google Scholar]
- Schwarzmann, F.; Jager, M.; Prang, N.; Wolf, H. The control of lytic replication of Epstein-Barr virus in B lymphocytes (Review). Int. J. Mol. Med. 1998, 1, 137–142. [Google Scholar]
- Hopwood, P.A.; Brooks, L.; Parratt, R.; Hunt, B.J.; Bokhari, M.; Thomas, J.A.; Yacoub, M.; Crawford, D.H. Persistent Epstein-Barr virus infection: Unrestricted latent and lytic viral gene expression in healthy immunosuppressed transplant recipients. Transplantation 2002, 74, 194–202. [Google Scholar] [CrossRef]
- Giot, J.F.; Mikaelian, I.; Buisson, M.; Manet, E.; Joab, I.; Nicolas, J.C.; Sergeant, A. Transcriptional interference between the EBV transcription factors EB1 and R: Both DNA-binding and activation domains of EB1 are required. Nucleic Acids Res. 1991, 19, 1251–1258. [Google Scholar] [CrossRef]
- Fixman, E.D.; Hayward, G.S.; Hayward, S.D. Trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J. Virol. 1992, 66, 5030–5039. [Google Scholar]
- Hislop, A.D.; Gudgeon, N.H.; Callan, M.F.; Fazou, C.; Hasegawa, H.; Salmon, M.; Rickinson, A.B. EBV-specific CD8+ T cell memory: Relationships between epitope specificity, cell phenotype, and immediate effector function. J. Immunol. 2001, 167, 2019–2029. [Google Scholar]
- Chang, L.K.; Chung, J.Y.; Hong, Y.R.; Ichimura, T.; Nakao, M.; Liu, S.T. Activation of Sp1-mediated transcription by Rta of Epstein-Barr virus via an interaction with MCAF1. Nucleic Acids Res. 2005, 33, 6528–6539. [Google Scholar] [CrossRef]
- Hsu, M.; Wu, S.Y.; Chang, S.S.; Su, I.J.; Tsai, C.H.; Lai, S.J.; Shiau, A.L.; Takada, K.; Chang, Y. Epstein-Barr virus lytic transactivator Zta enhances chemotactic activity through induction of interleukin-8 in nasopharyngeal carcinoma cells. J. Virol. 2008, 82, 3679–3688. [Google Scholar] [CrossRef]
- Tsai, S.C.; Lin, S.J.; Chen, P.W.; Luo, W.Y.; Yeh, T.H.; Wang, H.W.; Chen, C.J.; Tsai, C.H. EBV Zta protein induces the expression of interleukin-13, promoting the proliferation of EBV-infected B cells and lymphoblastoid cell lines. Blood 2009, 114, 109–118. [Google Scholar] [CrossRef]
- Ernberg, I.; Andersson, J. Acyclovir efficiently inhibits oropharyngeal excretion of Epstein-Barr virus in patients with acute infectious mononucleosis. J. Gen. Virol. 1986, 67, 2267–2272. [Google Scholar] [CrossRef]
- Funch, D.P.; Walker, A.M.; Schneider, G.; Ziyadeh, N.J.; Pescovitz, M.D. Ganciclovir and acyclovir reduce the risk of post-transplant lymphoproliferative disorder in renal transplant recipients. Am. J. Transplant. 2005, 5, 2894–2900. [Google Scholar] [CrossRef]
- Triantos, D.; Porter, S.R.; Scully, C.; Teo, C.G. Oral hairy leukoplakia: Clinicopathologic features, Pathogenesis, Diagnosis, and clinical significance. Clin. Infect. Dis. 1997, 25, 1392–1396. [Google Scholar]
- Meerbach, A.; Holy, A.; Wutzler, P.; de Clercq, E.; Neyts, J. Inhibitory effects of novel nucleoside and nucleotide analogues on Epstein-Barr virus replication. Antivir. Chem. Chemother. 1998, 9, 275–282. [Google Scholar]
- Chang, L.K.; Wei, T.T.; Chiu, Y.F.; Tung, C.P.; Chuang, J.Y.; Hung, S.K.; Li, C.; Liu, S.T. Inhibition of Epstein-Barr virus lytic cycle by (−)-epigallocatechin gallate. Biochem. Biophys. Res. Commun. 2003, 301, 1062–1068. [Google Scholar] [CrossRef]
- Yiu, C.Y.; Chen, S.Y.; Chang, L.K.; Chiu, Y.F.; Lin, T.P. Inhibitory effects of resveratrol on the Epstein-Barr virus lytic cycle. Molecules 2010, 15, 7115–7124. [Google Scholar] [CrossRef]
- Lin, T.P.; Chen, S.Y.; Duh, P.D.; Chang, L.K.; Liu, Y.N. Inhibition of the epstein-barr virus lytic cycle by andrographolide. Biol. Pharm. Bull. 2008, 31, 2018–2023. [Google Scholar] [CrossRef]
- Yi, T.; Zhang, H.; Cai, Z. Analysis of rhizoma polygoni cuspidati by HPLC and HPLC-ESI/MS. Phytochem. Anal. 2007, 18, 387–392. [Google Scholar] [CrossRef]
- Chu, X.; Sun, A.; Liu, R. Preparative isolation and purification of five compounds from the Chinese medicinal herb Polygonum cuspidatum Sieb. et Zucc by high-speed counter-current chromatography. J. Chromatogr. A 2005, 1097, 33–39. [Google Scholar] [CrossRef]
- Chang, J.S.; Liu, H.W.; Wang, K.C.; Chen, M.C.; Chiang, L.C.; Hua, Y.C.; Lin, C.C. Ethanol extract of Polygonum cuspidatum inhibits hepatitis B virus in a stable HBV-producing cell line. Antiviral Res. 2005, 66, 29–34. [Google Scholar] [CrossRef]
- Yiu, C.Y.; Chen, S.Y.; Huang, C.W.; Yeh, D.B.; Lin, T.P. Inhibitory effects of Polygonum cuspidatum on the epstein-barr virus lytic cycle. J. Food Drug Anal. 2011, 19, 107–113. [Google Scholar]
- Jones, R.J.; Seaman, W.T.; Feng, W.H.; Barlow, E.; Dickerson, S.; Delecluse, H.J.; Kenney, S.C. Roles of lytic viral infection and IL-6 in early versus late passage lymphoblastoid cell lines and EBV-associated lymphoproliferative disease. Int. J. Cancer 2007, 121, 1274–1281. [Google Scholar] [CrossRef]
- Hong, G.K.; Kumar, P.; Wang, L.; Damania, B.; Gulley, M.L.; Delecluse, H.J.; Polverini, P.J.; Kenney, S.C. Epstein-Barr virus lytic infection is required for efficient production of the angiogenesis factor vascular endothelial growth factor in lymphoblastoid cell lines. J. Virol. 2005, 79, 13984–13992. [Google Scholar] [CrossRef]
- Li, D.; Zhang, N.; Cao, Y.; Zhang, W.; Su, G.; Sun, Y.; Liu, Z.; Li, F.; Liang, D.; Liu, B.; et al. Emodin ameliorates lipopolysaccharide-induced mastitis in mice by inhibiting activation of NF-kappaB and MAPKs signal pathways. Eur. J. Pharmacol. 2013, 705, 79–85. [Google Scholar] [CrossRef]
- Lee, J.; Jung, E.; Lee, J.; Huh, S.; Hwang, C.H.; Lee, H.Y.; Kim, E.J.; Cheon, J.M.; Hyun, C.G.; Kim, Y.S.; et al. Emodin inhibits TNF alpha-induced MMP-1 expression through suppression of activator protein-1 (AP-1). Life Sci. 2006, 79, 2480–2485. [Google Scholar] [CrossRef]
- Chan, T.M.; Leung, J.K.; Tsang, R.C.; Liu, Z.H.; Li, L.S.; Yung, S. Emodin ameliorates glucose-induced matrix synthesis in human peritoneal mesothelial cells. Kidney Int. 2003, 64, 519–533. [Google Scholar] [CrossRef]
- Liu, P.; Liu, S.; Speck, S.H. Identification of a negative cis element within the ZII domain of the Epstein-Barr virus lytic switch BZLF1 gene promoter. J. Virol. 1998, 72, 8230–8239. [Google Scholar]
- Luka, J.; Kallin, B.; Klein, G. Induction of the Epstein-Barr virus (EBV) cycle in latently infected cells by n-butyrate. Virology 1979, 94, 228–231. [Google Scholar] [CrossRef]
- Carmichael, J.; DeGraff, W.G.; Gazdar, A.F.; Minna, J.D.; Mitchell, J.B. Evaluation of a tetrazolium-based semiautomated colorimetric assay: Assessment of radiosensitivity. Cancer Res. 1987, 47, 943–946. [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Sample Availability: Not available.
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yiu, C.-Y.; Chen, S.-Y.; Yang, T.-H.; Chang, C.-J.; Yeh, D.-B.; Chen, Y.-J.; Lin, T.-P. Inhibition of Epstein-Barr Virus Lytic Cycle by an Ethyl Acetate Subfraction Separated from Polygonum cuspidatum Root and Its Major Component, Emodin. Molecules 2014, 19, 1258-1272. https://doi.org/10.3390/molecules19011258
Yiu C-Y, Chen S-Y, Yang T-H, Chang C-J, Yeh D-B, Chen Y-J, Lin T-P. Inhibition of Epstein-Barr Virus Lytic Cycle by an Ethyl Acetate Subfraction Separated from Polygonum cuspidatum Root and Its Major Component, Emodin. Molecules. 2014; 19(1):1258-1272. https://doi.org/10.3390/molecules19011258
Chicago/Turabian StyleYiu, Ching-Yi, Shih-Ying Chen, Tsai-Hsiu Yang, Che-Jung Chang, Dong-Bor Yeh, Yi-Jie Chen, and Tsuey-Pin Lin. 2014. "Inhibition of Epstein-Barr Virus Lytic Cycle by an Ethyl Acetate Subfraction Separated from Polygonum cuspidatum Root and Its Major Component, Emodin" Molecules 19, no. 1: 1258-1272. https://doi.org/10.3390/molecules19011258



