Biotransformation of Isoflavone Using Enzymatic Reactions
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemical Structure of 7,3',4'-Trihydroxyisoflavone from Daidzein

2.2. Identification and Functional Construction of CYP105D7 and Redox Partner in E. coli System

2.3. Overexpression and Purification of CYP105D7, PdR and Pdx

2.4. Characterization of Recombinant CYP105D7 and Kinetic Parameter
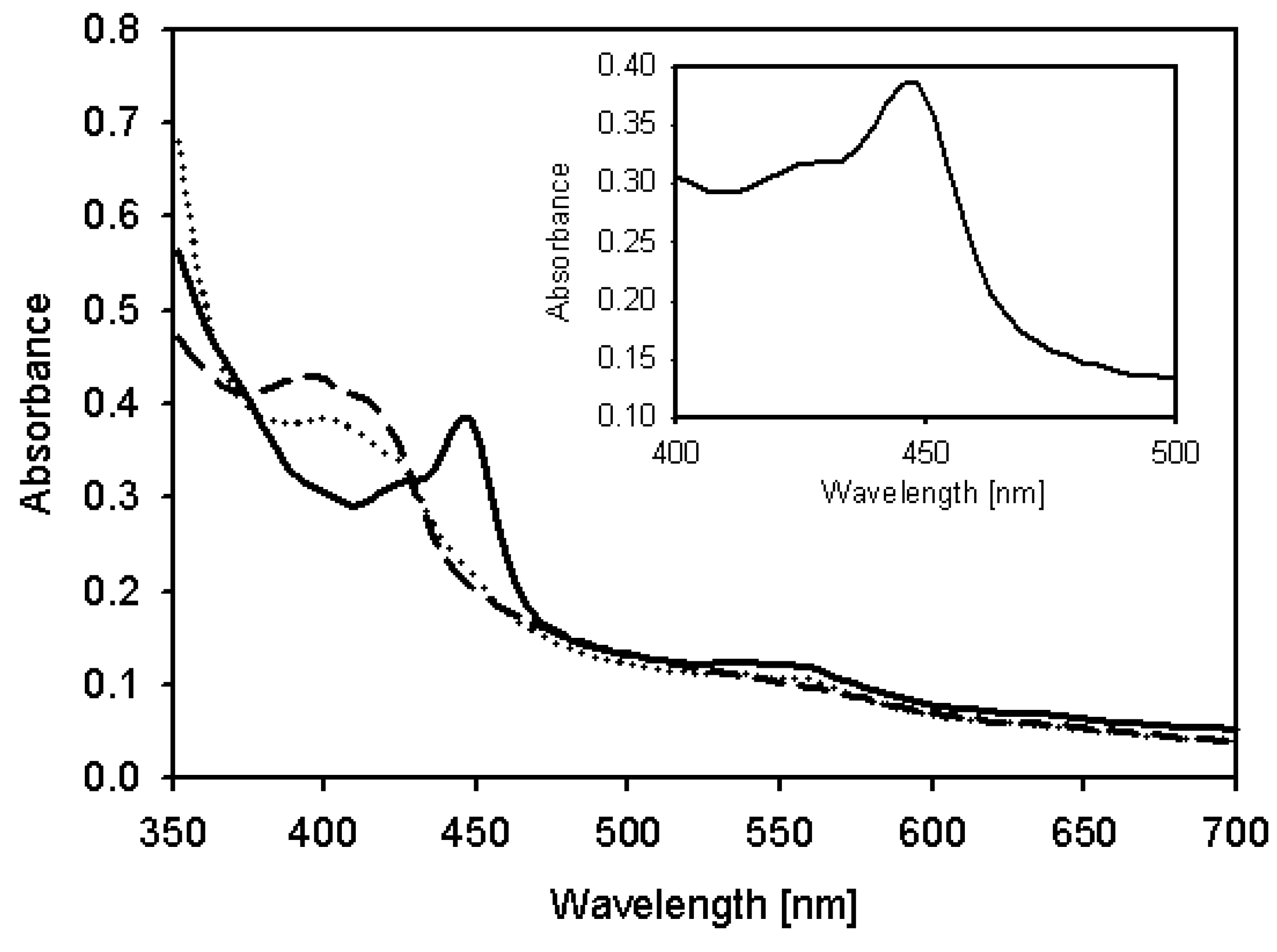
| Substrate | Km [μM] | kcat [min−1] | kcat/Km [μM−1/min−1] * |
|---|---|---|---|
| Daidzein | 21.83 ± 6.3 | 15.01 ± 0.6 | 0.69 |
2.5. Hydroxylation of Daidzein by CYP105D7 in Vivo and in Vitro System

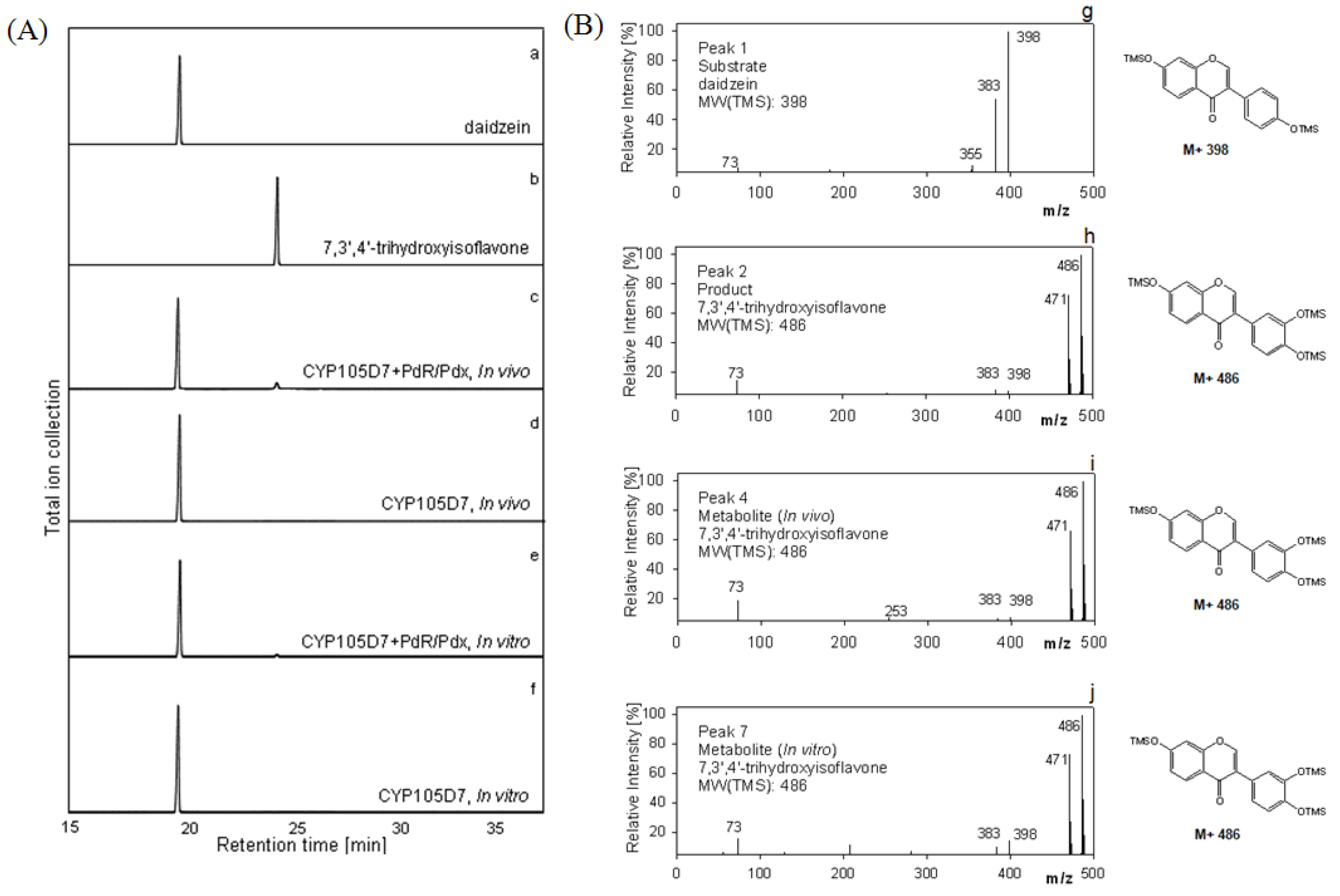
2.6. Effect of P450 Inhibitors on the Hydroxylation of Daidzein
| P450 inhibitor * | Reactive activity (%) |
|---|---|
| None | 100 |
| Coumarin | 2 |
| Erythromycin | 63 |
| Ketoconazole | 14 |
3. Experimental
3.1. Chemicals
3.2. Plasmid Construction
3.3. Cloning of CYP105D7 from Streptomyces avermitilis MA4680
3.4. Coexpression of CYP105D7 and Redox Partner for in Vivo System
3.5. Purification of CYP105D7 and Redox Partner for in Vitro System
3.6. CYP105D7 Concentration Measurement and Absorption Spectra
3.7. Kinetic Study for Substrate Activity with CYP105D7
3.8. Daidzein Hydroxylation in Vivo and in Vitro System
3.9. High Performance Liquid Chromatography (HPLC) Analysis of Product
3.10. Gas Chromatography (GC)/Mass Spectrometry (MS) Analysis
3.11. Effects of P450 Inhibitors on Hydroxylation of Daidzein
4. Conclusions
Acknowledgments
References
- Klus, K.; Barz, W. Formation of polyhydroxylated isoflavones from the soybean seed isoflavones daidzein and glycitein by bacteria isolated from tempe. Arch. Microbiol. 1995, 164, 428–434. [Google Scholar] [CrossRef]
- Esaki, H.; Watanabe, R.; Osaka, T. Formation mechanism for potent antioxidative o-dihydroxyisoflavones in soybeans fermented with Aspergillus saitori. Biosci. Biotechnol. Biochem. 2006, 63, 851–858. [Google Scholar] [CrossRef]
- Kulling, S.E.; Honig, D.M.; Simat, T.J.; Metzler, M. Oxidative in vitro metabolism of the soy phytoestrogens daidzein and genistein. J. Agric. Food. Chem. 2000, 48, 4963–4972. [Google Scholar] [CrossRef]
- Kulling, S.E.; Honig, D.M.; Metzler, M. Oxidative metabolism of the soy isoflavones daidzein and genistein in humans in vitro and in vivo. J. Agric. Food. Chem. 2001, 49, 3024–3033. [Google Scholar] [CrossRef]
- Hirota, A.; Inaba, M.; Chen, Y.C.; Abe, N.; Taki, S.; Yano, M.; Kawaii, S. Isolation of 8-hydroxyglycitein and 6-hydroxydaidzein from soybean miso. Biosci. Biotechnol. Biochem. 2004, 68, 1372–1374. [Google Scholar] [CrossRef]
- Rufer, C.E.; Kulling, S.E. Antioxidant activity of isoflavones and their major metabolites using different in vitro assays. J. Agric. Food. Chem. 2006, 54, 2926–2931. [Google Scholar] [CrossRef]
- Rufer, C.E.; Maul, R.; Donauer, E.; Fabian, E.J.; Kulling, S.E. In vitro and in vivo metabolism of the soy isoflavone glycitein. Mol. Nutr. Food Res. 2007, 51, 813–823. [Google Scholar] [CrossRef]
- Chang, T.S.; Ding, H.Y.; Lin, H.C. Identifying 6,7,4'-trihydroxyisoflavone as a Potent Tyrosinase Inhibitor. Biosci. Biotech. Biochem. 2005, 69, 1999–2001. [Google Scholar] [CrossRef]
- Akiyama, T.; Ishida, J.; Nakagawa, S.; Ogawara, H.; Watanabe, S.; Itoh, N.; Shibuya, M.; Fukami, Y. Genistein, a specific inhibitor of tyrosine specific protein kinases. J. Biol. Chem. 1987, 262, 5592–5595. [Google Scholar]
- Rufer, C.E.; Glatt, H.; Kulling, S.E. Structural elucidation of hydroxylated metabolites of the isoflavane equol by gas chromatography-mass spectrometry and high-performance liquid chromatography-mass spectrometry. Drug. Metab. Dispos. 2006, 34, 51–60. [Google Scholar] [CrossRef]
- Ortiz de Montellano, P.R. Cytochrome P450: Structure, Mechanism, and Biochemistry, 3rd ed.; Kluwer Academic/Plenum Publishers: New York, NY, USA.
- Bernhardt, R. Cytochromes P450 as versatile biocatalysts. J. Biotechnol. 2006, 124, 128–145. [Google Scholar] [CrossRef]
- Urlacher, V.; Schmid, R.D. Biotransformations using prokaryotic P450 monooxygenases. Curr. Opin. Biotechnol. 2002, 13, 557–564. [Google Scholar] [CrossRef]
- Holland, H.L.; Weber, H.K. Enzymatic hydroxylation reactions. Curr. Opin. Biotechnol. 2000, 11, 547–553. [Google Scholar] [CrossRef]
- Taylor, M.; Lamb, D.C.; Cannell, R.; Dawson, M.; Kelly, S.L. Cytochrome P450105D1 (CYP105D1) from Streptomyces griseus: Heterologous expression, activity, and activation effects of multiple xenobiotics. Biochem. Biophys. Res. Commun. 1999, 263, 838–842. [Google Scholar] [CrossRef]
- Staunton, J.; Weissman, K.J. Polyketide biosynthesis: A millennium review. Nat. Prod. Rep. 2001, 18, 380–416. [Google Scholar] [CrossRef]
- Shen, B. Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Curr. Opin. Chem. Biol. 2003, 7, 285–295. [Google Scholar] [CrossRef]
- Lee, K.W.; Joo, H.S.; Yang, Y.H.; Song, E.J.; Kim, B.G. Proteomics for Streptomyces: “Industrial Proteomics” for Antibiotics. J. Microbiol. Biotechnol. 2006, 16, 331–348. [Google Scholar]
- Ikeda, H.; Ishikawa, J.; Hanamoto, A.; Shinose, M.; Kikuchi, H.; Shiba, T.; Sakaki, Y.; Hattori, M.; Omura, S. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 2003, 21, 526–531. [Google Scholar] [CrossRef]
- Liu, W.; Rosazza, J.P. A soluble Bacillus cereus cytochrome P-450cin system catalyzes 1,4-cineole hydroxylations. Appl. Environ. Microbiol. 1993, 59, 3889–3893. [Google Scholar]
- Orita, M.; Yamamoto, S.; Katayama, N.; Aoki, M.; Takayama, K.; Yamagiwa, Y.; Seki, N.; Suzuki, H.; Kurihara, H.; Sakashita, H.; et al. Coumarin and Chromen-4-one analogues as tautomerase inhibitors of macrophage migration inhibitory factor: Discovery and X-ray Crystallography. J. Med. Chem. 2001, 44, 540–547. [Google Scholar] [CrossRef]
- Harris, R.; Wood, D.M.; Bottomley, L.; Blagg, S.; Owen, K.; Hughes, P.; Waring, R.H. Phytoestrogens are potent inhibitors of estrogen sulfation: Implications for breast cancer risk and treatment. J. Clin. Endocrinol. Metab. 2004, 89, 1779–1787. [Google Scholar] [CrossRef]
- Bradford, M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Omura, T.; Sato, R.J. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J. Biol. Chem. 1964, 239, 2370–2378. [Google Scholar]
- Gunsalus, I.C.; Wagner, G.C. [17] Bacterial p450cam methylene monooxygenase components: Cytochrome m, putidaredoxin, and putidaredoxin reductase. Methods Enzymol. 1978, 52, 166–188. [Google Scholar] [CrossRef]
- Sample Availability: Not available.
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Roh, C. Biotransformation of Isoflavone Using Enzymatic Reactions. Molecules 2013, 18, 3028-3040. https://doi.org/10.3390/molecules18033028
Roh C. Biotransformation of Isoflavone Using Enzymatic Reactions. Molecules. 2013; 18(3):3028-3040. https://doi.org/10.3390/molecules18033028
Chicago/Turabian StyleRoh, Changhyun. 2013. "Biotransformation of Isoflavone Using Enzymatic Reactions" Molecules 18, no. 3: 3028-3040. https://doi.org/10.3390/molecules18033028




