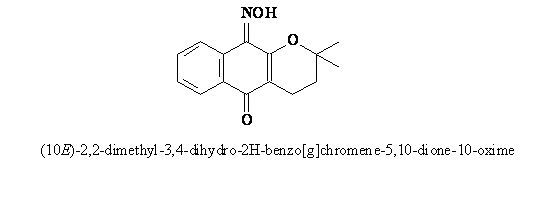
Crystal Structure and Hydrogen Bonding Study of (10E)-2,2-Dimethyl-3,4-dihydro-2H-benzo[g]chromene-5,10-dione 10-Oxime Derived From a-Lapachone
Abstract
:1. Introduction
2. Results and Discussion

2.1. Infrared, 1H- and 13C-Nuclear Magnetic Resonance Spectroscopy
| Carbon Atom | δ 13C | δ 1H ( J in Hz ) |
|---|---|---|
| 1 | 184.0 | - |
| 2 | 113.6 | - |
| 3 | 156.6 | - |
| 4 | 139.7 | - |
| 5 | 126.9 | - |
| 6 | 129.8 | 9.06 (d, 7.8) |
| 7 | 132.6 | 7.65 (td, 1.4, 7.6) |
| 8 | 130.7 | 7,58 (td, 1.2, 7.7) |
| 9 | 126.8 | 8.27 (dd, 1.2, 7.6) |
| 10 | 130.7 | - |
| 11 | 16.9 | 2.63 (t, 6.6) |
| 12 | 31.8 | 1.88 (t, 6.6) |
| 13 | 78.4 | - |
| 14 | 26.8 | 1.48 (s) |
| 15 | 26.8 | 1.48 (s) |
| 16 | - | 12.25 (s) |
2.2. Crystal Structure

| Bonds | |
| N1-O1 | 1.382(1) |
| N1-C4 | 1.290(2) |
| C1-O2 | 1.234(2) |
| C3-O3 | 1.353(2) |
| C13-O3 | 1.473(2) |
| Angles | |
| O1-N1-C4 | 117.2(1) |
| C3-O3-C13 | 118.1(1) |
| O2-C1-C10 | 120.8(1) |
| O2-C1-C2 | 120.6(1) |
| C12-C13-C15 | 111.8(1) |
| O3-C13-C14 | 107.8(1) |


| D-H···A | D-H | H··· A | D···A |  D-H···A D-H···A |
|---|---|---|---|---|
| O1-H16...O3 i | 0.86(2) | 2.35(2) | 3.070(2) | 142(2) |
| O1-H16...N1 i | 0.86(2) | 2.01(2) | 2.771(2) | 149(2) |
| C6-H6…O1 | 0.93 | 2.16 | 2.785(2) | 123 |
| D-H···Cg | D-H | H··· Cg | D···Cg |  D-H···Cg D-H···Cg |
| C15-H15c…Cgii | 1.02(2) | 2.795 | 3.797 | 160.28 |
3. Experimental
3.1. General
3.2. Synthesis of 1
3.3. X-ray diffraction Experiment
| Empirical formula | C15H15NO3 |
| Formula weight | 257.28 |
| Temperature | 295(2) K |
| Wavelength | 0.71073 A |
| Crystal system, space group | triclinic, P-1 |
| Unit cell dimensions | a = 6.6069(13) Å α = 91.47(3)° |
| b = 9.6001(19) Å β = 94.47(3)° | |
| c = 10.176(2) Å γ = 94.27(3)° | |
| Volume | 641.3(2) Å3 |
| Z, Calculated density | 2, 1.332 mg/m3 |
| Absorption coefficient | 0.093 mm-1 |
| F(000) | 272 |
| Crystal size | 0.47 × 0.40 × 0.20 mm |
| Theta range for data collection | 2.97 to 25.00° |
| Limiting indices | −7<=h<=7, −11<=k<=11, −12<=l<=12 |
| Reflections collected / unique | 10469 / 2246 [R(int) = 0.0339] |
| Completeness to theta = 25.00 | 99.5 % |
| Max. and min. transmission | 0.9816 and 0.9574 |
| Refinement method | Full-matrix least-squares on F2 |
| Data / restraints / parameters | 2246 / 0 / 216 |
| Goodness-of-fit on F2 | 1.032 |
| Final R indices [I>2sigma(I)] | R1 = 0.0390, wR2 = 0.1010 |
| R indices (all data) | R1 = 0.0550, wR2 = 0.1103 |
| Largest diff. peak and hole | 0.137 and −0.190 e.Å-3 |
4. Conclusions
Acknowledgements
Supplementary Material
References and Notes
- Alcalde, E.; Mesquida, N.; Alvarez-Rúa, C.; Cuberes, R.; Frigola, J.; García-Granda, S. 1,2-Diaryl(3-pyridyl)ethanone Oximes. Intermolecular Hydrogen Bonding Networks Revealed by X-ray Diffraction. Molecules 2008, 13, 301–318. [Google Scholar] [CrossRef]
- Pérez-Sacau, E.; Estévez-Braum, A.; Ravelo, A.G.; Ferro, E.A.; Tokuda, H.; Mukainaka, T.; Nishino, H. Inhibitory effects of lapachol derivatives on Epstein-Barr virus activation. Bioorg. Med. Chem. 2003, 11, 483–488. [Google Scholar] [CrossRef]
- Pérez-Sacau, E.; Diaz-Peñate, R.G.; Estévez-Braun, A.; Ravelo, A.G.; García-Castellano, J.M.; Pardo, L.; Campillo, M. Synthesis and Pharmacophore Modeling of Naphthoquinone Derivatives with Cytotoxic Activity in Human Promyelocytic Leukemia HL-60 Cell Line. J. Med. Chem. 2007, 50, 696–706. [Google Scholar]
- Lemos, T.L.G.; Monte, F.J.Q.; Santos, A.K.L.; Fonseca, A.M.; Santos, H.S.; Oliveira, M.F.; Costa, S.M.O.; Pessoa, O.D.L.; Braz-Filho, R. Quinones from plants of Northeastern Brazil: Structural diversity, chemical transformations, NMR data and biological activities. Nat. Prod. Res. 2007, 21, 529–550. [Google Scholar] [CrossRef]
- Burnett, A.R.; Thomson, R.H. Naturally occurring quinones. Part X. The quinonoid constituents of Tabebuia avellanedae (Bignoniaceae). J. Chem. Soc. C 1967, 2100–2104. [Google Scholar]
- Hussain, H.; Krohn, K.; Ahmad, V.U.; Miana, G.A.; Green, I.R. Lapachol: an overview. ARKIVOC 2007, ii, 145–171. [Google Scholar]
- Hooker, S.C. LVII.—The constitution of “lapachic acid” (lapachol) and its derivatives. J. Chem. Soc. Trans. 1892, 61, 611–650. [Google Scholar] [CrossRef]
- Pinto, A.V.; Pinto, M.C.F.R.; Oliveira, C.G.T. Synthesis of the α- and nor-β-lapachones. Properties in acid and reactions with N-bromosuccinimide. An. Acad. Bras. Cienc. 1982, 54, 107–114. [Google Scholar]
- Yu, H. l. 6-Hydroxy-3-(hydroxyimino)indolin-2-one. Acta Crystallogr. 2009, E65, o2328. [Google Scholar]
- Yuan, X.-Y.; Zhang, M.; Ng, S.W. Perillartine. Acta Crystallogr. 2009, E65, o2149. [Google Scholar]
- Abbas, A.; Hussain, S.; Hafeez, N.; Badshah, A.; Hasan, A.; Lo, K.M. (E)-4-Nitrobenzaldehyde oxime. Acta Crystallogr. 2010, E66, o1130. [Google Scholar]
- Allen, F.H.; Watson, D.G.; Brammer, L.; Orpen, A.G.; Taylor, R. Typical Interatomic Distances: Organic Compounds. In International Tables for Crystallography; Prince, E., Ed.; Wiley: New York, NY, USA, 2006; Volume C, pp. 790–811. Chapter 9.5. [Google Scholar]
- Spek, A.L. Single-crystal structure validation with the program PLATON. J. Appl. Crystallogr. 2003, 36, 7–13. [Google Scholar] [CrossRef]
- Jeffrey, G.A.; Saenger, W. Hydrogen Bonding in Biological Structures; Springer-Verlag: Berlin, Germany, 1994; p. 569. [Google Scholar]
- Hörner, M.; Oliveira, G.M.; Visentin, L.C.; Cezar, R. Dimerization and polymeric self arrangement of [HgII(Py)2(PhN2C6H4NNNC6H4R)2] (Py = pyridine; R = NO2) through reciprocal metal-η2-arene π-interactions and non-classical C–H…O bonding: Synthesis and X-ray characterization of a polyaryl asymmetric-substituted triazenide complex polymer of Hg(II). Inorg. Chim. Acta 2006, 359, 4667–4671. [Google Scholar] [CrossRef]
- Haiduc, I.; Edelmann, F.T. Supramolecular Organometallic Chemistry; Wiley–VCH: Weinheim, Germany, 1999; p. 486. [Google Scholar]
- Simard, S.; Su, D.; Wuest, J.D. Use of hydrogen bonds to control molecular aggregation. Self-assembly of three-dimensional networks with large chambers. J. Am. Chem. Soc. 1991, 113, 4696–4698. [Google Scholar] [CrossRef]
- Fyfe, M.C.T.; Stoddart, J.F. Synthetic Supramolecular Chemistry. Acc. Chem. Res. 1997, 30, 393–401. [Google Scholar] [CrossRef]
- Farrugia, L.J. ORTEP-3 for Windows - a version of ORTEP-III with a Graphical User Interface (GUI). J. Appl. Crystallogr. 1997, 30, 565–566. [Google Scholar] [CrossRef]
- Hooft, R.W.W. Collect; Nonius BV: Delft, The Netherlands, 1998. [Google Scholar]
- Duisenberg, A.J.M.; Hooft, R.W.W.; Schreurs, A.M.M.; Kroon, J. Accurate cells from area-detector images. J. Appl. Crystallogr. 2000, 33, 893–898. [Google Scholar] [CrossRef]
- Duisenberg, A.J.M. Indexing in single-crystal diffractometry with an obstinate list of reflections. J. Appl. Crystallogr. 1992, 25, 92–96. [Google Scholar] [CrossRef]
- Duisenberg, A.J.M.; Kroon-Batenburg, L.M.J.; Schreurs, A.M.M. An intensity evaluation method: EVAL-14. J. Appl. Crystallogr. 2003, 36, 220–229. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SADABS, Program for Empirical Absorption Correction of Area Detector Data; University of Göttingen: Germany, 1996. [Google Scholar]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. 2008, A64, 112–122. [Google Scholar]
- Sample Availability: Samples of compound 1 are available from the authors.
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Da Silva, A.R.; Herbst, M.H.; Ferreira, A.B.B.; Da Silva, A.M.; Visentin, L.C. Crystal Structure and Hydrogen Bonding Study of (10E)-2,2-Dimethyl-3,4-dihydro-2H-benzo[g]chromene-5,10-dione 10-Oxime Derived From a-Lapachone. Molecules 2011, 16, 1192-1200. https://doi.org/10.3390/molecules16021192
Da Silva AR, Herbst MH, Ferreira ABB, Da Silva AM, Visentin LC. Crystal Structure and Hydrogen Bonding Study of (10E)-2,2-Dimethyl-3,4-dihydro-2H-benzo[g]chromene-5,10-dione 10-Oxime Derived From a-Lapachone. Molecules. 2011; 16(2):1192-1200. https://doi.org/10.3390/molecules16021192
Chicago/Turabian StyleDa Silva, Andrea R., Marcelo H. Herbst, Aurelio B. B. Ferreira, Ari M. Da Silva, and Lorenzo C. Visentin. 2011. "Crystal Structure and Hydrogen Bonding Study of (10E)-2,2-Dimethyl-3,4-dihydro-2H-benzo[g]chromene-5,10-dione 10-Oxime Derived From a-Lapachone" Molecules 16, no. 2: 1192-1200. https://doi.org/10.3390/molecules16021192