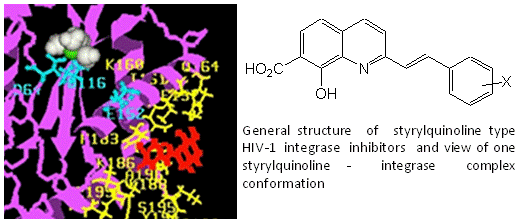
Chemistry and Structure-Activity Relationship of the Styrylquinoline-Type HIV Integrase Inhibitors
Abstract
:Abbreviations
| HIV | Human immunodeficiency virus |
| IN | Integrase |
| INSTI | integrase strand transfer inhibitor |
| INBI | integrase binding inhibitor |
| SQL | styrylquinoline |
| LTR | long terminal repeat |
| DMAP | 4-Dimethylaminopyridine |
| DCC | Dicyclohexylcarbodiimide |
| DMF | Dimethylformamide |
| TFA | Trifluoroacetic acid |
| DFT | Density functional theory |
| RSV | Rous sarcoma virus |
| CoMFA | Comparative Molecular Field Analysis |
| QSAR | Quantitative structure-activity relationship |
| AZT | Azidothymidine. |
1. Introduction

2. Chemistry and Structure-Activity Relationship of Styrylquinolines in vitro

2.1. Discovery of the 8-hydroxy-quinoline-7-carboxylic acid- pharmacophore

| Compound | 3’ProcessingIC50 (μM) | Strand Transfer IC50 (μM) | Reference | |
|---|---|---|---|---|
| 2b |  | >100.000 | >100.000 | 14 |
| 3a |  | >100.000 | >100.000 | 19 |
| 3b |  | >100.000 | 114.000 | 14 |
| 4 |  | 77.000 | untested | 19 |
| 7 |  | >100.000 | untested | 19 |
| 12 |  | >100.000 | untested | 21 |
| 10 |  | 47.000 | untested | 21 |
| 5 |  | >100.000 | untested | 20 |
| 13 |  | 0.033 | 23 | |
| 14 |  | 0.370 | 22 | |
2.2.Modulation of the C-8 substituent of the styrylquinoline inhibitors

| Compd.Nb |  | 3’-Processing IC50 (μM) | References | ||||
|---|---|---|---|---|---|---|---|
| R5 | R6 | R7 | R8 | X | |||
| 21 | H | H | H | H | CH | >100.0 | 14 |
| 22 | H | H | H | OH | CH | 7.4 | 14 |
| 20 | H | H | H | OH | N | 57.0 | 24 |
| 23 | H | H | H | OAc | CH | >100.0 | 14 |
| 24 | H | H | H | NO2 | CH | >100.0 | 14 |
| 25 | H | H | H | NH2 | CH | >100.0 | 14 |
| 26 | H | H | H | CO2H | CH | >100.0 | 25 |
| 27 | CO2H | H | H | H | CH | >100.0 | 25 |
| 28 | H | CO2H | H | H | CH | >100.0 | 25 |
| 29 | H | H | CO2H | H | CH | >100.0 | 25 |
| 30 | CO2H | H | CO2H | H | CH | >100.0 | 25 |
| 31 | OH | CO2H | H | H | CH | >100.0 | 19 |
| 32 | CO2H | H | H | OH | CH | 7.0 | 19 |
2.3.Modulation of the C-7 substituent on the 8,3',4'-trihydroxystyrylquinoline scaffold



 | 3’-Processing IC50 (μM) | Strand Transfer IC50 (μM) | Reference | |
|---|---|---|---|---|
| 34 | X = CN | 3.0 | Untested | 14 |
| 35 | X = CO2H | 2.4 | 1 | 14 |
| 54 | X = CO2CH3 | >100 | >100 | 26 |
| 42 |  | 2a | 30 | |
| 43 |  | Untested | 2.4 | 30 |
| 46 | X = (HO)2PO | Untested | >100 | 30 |
| 47 | X = HO2CCO | 100 | >100 | 32 |
| 53 |  | 2.3 | Untested | 14 |
| 56 |  | 100 | Untested | 30 |
| 55 |  | >300 | Untested | 19 |

 | 3’-Processing IC50 (μM) | Strand Transfer IC50 (μM) | References |
|---|---|---|---|
| 59a, R = H | 82.0 | Untested | 30 |
| 59b, R = Ph | 50.0a | Untested | 33 |
| 59c, R = 1-Naphthyl | 92.0 | Untested | 33 |
| 59da, R = CH3 | 0.2 | 0.2 | 34 |
| 59e, R = C8H17 | >100.0 b | Untested | 33 |
| 59f, R = PhCH2CH2 | >100.0 a | Untested | 33 |
| 59g, R = 4-NO2Ph | 100.0 | >100.0 | 33 |
| 59h, R = 2-NO2Ph | >100.0 | 70.0 | 33 |
| 59i, R = 3,4-F2Ph- | 24.0 | >100.0 | 33 |
| 59j, R = 4-pyridyl | >100.0 a | Untested | 33 |
| 59k, R = 3-pyridyl | >100.0 | >100.0 | 33 |
| 59l, R = 2-OH-Ph | >100.0 | >100.0 | 33 |
| 5m, R = 4-OH-Ph | 65.0 | >100.0 | 33 |
| 59n, R = 4-(HO2C)Ph | 0.2 | Untested | 33 |
| 59o, R = 3-(HO2C)Ph | 0.2 | Untested | 33 |
| 59p, R = 2-(HO2C)Ph | 0.3 | Untested | 33 |
2.4. Functionalization of the C-5 quinoline position


| Compd. Numb. |  | In vitro biological activity | Reference | ||
|---|---|---|---|---|---|
| R5 | R7 | 3’-Processing IC50 (μM) | Strand TransferIC50 (μM) | ||
| 60 | Cl | Cl | >100.00 | >100.00 | 26 |
| 61 | Br | Br | 21.00 | 33.00 | 19 |
| 62 | Br | CO2H | 5.00 | Untested | 30 |
| 63a | CO2H | CO2H | 0.20 | 0.07 | 34 |
| 64 |  | CO2H | 1.40 | 1.00 | 34 |
| 65 |  | CO2H | >100.00c | Untested | 19 |
| 76b | Cl | SO2NHpClPh | >100.00 | Untested | 36 |
2.5. Modulation of the ancillary ring
 | In vitro Biological activity | Reference | ||
|---|---|---|---|---|
| Compd. nb. | R | 3’-Processing IC50(μM) | Strand Transfer IC50 (μM) | |
| 77a |  | 5.3 | 2.1 | 14 |
| 77b |  | 1.9 | 5.1 | 26 |
| 77c |  | 3.4 | 3.0 | 26 |
| 77d |  | 4.0 | 11.0 | 26 |
| 77e |  | 1.6 | untested | 26 |
| 77f |  | 2.2 | 3.5 | 26 |
| 77g |  | 1.2 | 1.7 | 26 |
| 77h |  | 3.4 | 31.0 | 26 |
| 77i |  | 5.0 | untested | 25 |
| 77j |  | 39.0 | 30.0 | 30 |
| 77k |  | >100.0 | untested | 30 |
 | In vitro Biological activity | Reference | ||
|---|---|---|---|---|
| Compd. nb. | R | 3’-Processing IC50(μM) | Strand Transfer IC50 (μM) | |
| 35 |  | 2.40 | 1.00 | 14 |
| 77l |  | 3.20 | 3.20 | 26 |
| 77m |  | 3.70 | 2.80 | 14 |
| 77n |  | 0.60 | 0.03 | 26 |
| 77o |  | 10.00 | 30.00 | 25 |
| 77p |  | 5.00 | untested | 25 |
| 77q |  | 2.80 | 3.70 | 26 |
| 77r |  | 1.30 | 3.00 | 26 |
| 77s |  | 2.70 | 0.60 | 14 |
| 77t |  | 0.30 | 0.40 | 14 |
| 77u |  | 0.70 | 2.80 | 26 |
| 77v |  | 5.40 | untested | 26 |
| 77w |  | 4.90 | 4.50 | 26 |
| 77x |  | 0.30 | 0.30 | 37 |
| 77y |  | 1.20 | 1.00 | 26 |
| 77z |  | 2.00 | 2.00 | 19 |
| 77aa |  | 7.00 | 4.00 | 19 |
| 77ab |  | 7.00 | 7.00 | 19 |
| 77ac |  | 10.00 | untested | 19 |
2.6. Modulation of the spacer


 | In vitro biological activity | Reference | ||
|---|---|---|---|---|
| Compd nb. | X | 3’-Processing IC50(μM) | Strand Transfer IC50 (μM) | |
| 35 | HC=CH | 2.4 | 1.0 | 14 |
| 78 | H2CCH2 | 2.3 | 1.5 | 26 |
| 92 | CONH | 0.9 | untested | 18 |
| 93 | CONHCH2 | 5.0 | 18 | |
| 94 | HNCONHCH2 | >100.0 | 18 | |
| 95 | HNCONH | >100.0 | 18 | |
3. Physico-Chemical Rational of the Styrylquinoline-Integrase Interaction
3.1. Magnesium binding ability of styrylquinoline integrase inhibitors
 | In vitro biological activity | ||
|---|---|---|---|
| Compd. Nb. | R | 3’-Processing, IC50 (μM) | 3’-Processing activity of the corresponding SQL IC50 (μM) |
| 96 |  | 6.5 | 0.7 (77u) |
| 97 |  | 5.0 | 0.6 (77n) |
| 98 |  | 5.0 | 3.7 (77m) |
| 99 |  | >100.0 | 3.2 (77l) |
| 100 |  | 6.5 | 0.7 (77u) |
| 101 |  | 1.5 | _ |
| 102 |  | >100.0 | 5.3 (77a) |
| 103 |  | >100.0 | 5.3 (77a) |
| 104 |  | 3.0 | _ |
| 105 |  | >100.0 | _ |
| 106 |  | 2.9 | _ |

3.2. Comparative molecular field analysis and docking studies

5. Conclusions
References
- Lalezari, J.P.; Henry, K.; O'Hearn, M.; Montaner, J.S.; Piliero, P.J.; Trottier, B.; Walmsley, S.; Cohen, C.; Kuritzkes, D.R.; Eron, J.J., Jr.; Chung, J.; DeMasi, R.; Donatacci, L.; Drobnes, C.; Delehanty, J.; Salgo, M. Enfuvirtide, an HIV-1 fusion inhibitor, for drug-resistant HIV infection in North and South America. N. Engl. J. Med. 2003, 348, 2175–2185. [Google Scholar]
- Stellbrink, H.-J. Novel compounds for the treatment of HIV type-1 infection. Antivir. Chem. Chemother. 2009, 19, 189–200. [Google Scholar]
- Hunt, J.S.; Romanelli, F. Maraviroc, a CCR5 coreceptor antagonist that blocks entry of human immunodeficiency virus type 1. Pharmacotherapy 2009, 29, 295–304. [Google Scholar]
- Pommier, Y.; Johnson, A.A.; Marchand, C. Integrase inhibitor to treat HIV/AIDS. Nature Rev. Drug Disc. 2005, 4, 236–247. [Google Scholar]
- Lewinski, M.K.; Bushman, F.D. Retroviral DNA integration-mechanism and consequences. Adv. Genet. 2005, 55, 147–181. [Google Scholar]
- Chirch, L.M.; Morrison, S.; Steigbigel, R.T. Treatment of HIV infection with raltegravir. Expert Opin. Pharmaco. 2009, 10, 1203–1211. [Google Scholar]
- Vandeckerckhove, L. GSK-1349572, a novel integrase inhibitor for the treatment of HIV infection. Curr. Opin. Investig. Drugs 2010, 11, 203–212. [Google Scholar]
- Katlama, C.; Murphy, R. Emerging role of integrase inhibitors in the management of treatment-experienced patients with HIV infection. Ther. Clin. Risk Manag. 2009, 5, 331–340. [Google Scholar]
- Beck-Engeser, G.B.; Eilat, D.; Harrer, D.; Jäck, H.-M.; Wabl, M. Early onset of autoimmune disease by the retroviral integrase inhibitor raltegravir. Proc. Natl. Acad. Sci. USA 2009, 106, 20865–20870. [Google Scholar]
- Williams, N.H.; Takasaki, B.; Wall, M.; Chin, J. Structure and nuclease activity of simple dinuclear metal complexes: Quantitative dissection of the role of metal ions. Acc. Chem. Res. 1999, 32, 485–493. [Google Scholar]
- Beese, L.S.; Steitz, T.A. Structural basis for the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991, 10, 25–33. [Google Scholar]
- Goldgur, Y.; Dyda, F.; Hickman, A.B.; Jenkins, T.M.; Craigie, R.; Davies, D.R. Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium. Proc. Natl. Acad. Sci. USA 1998, 95, 9150–9154. [Google Scholar]
- Lins, R.D.; Adesokan, A.; Soares, T.A.; Briggs, J.M. Investigations on human immunodeficiency virus type 1 integrase/DNA binding interactions via molecular dynamics and electrostatics calculations. Pharmacol. Therapeut. 2000, 85, 123–131. [Google Scholar]
- Mekouar, K.; Mouscadet, J.-F.; Desmaële, D.; Subra, F.; Savouré, D.; Auclair, C.; d'Angelo, J. Styrylquinoline derivatives: a new class of potent HIV-1-integrase inhibitors that block HIV-1 replication in CEM cells. J. Med. Chem. 1998, 41, 2846–2857. [Google Scholar]
- Meek, W.H.; Fuschman, C.H. Carboxylation of substituted phenol in N,N-dimethylamide solvents at atmospheric pressure. J. Chem. Eng. Data 1969, 14, 388–391. [Google Scholar] [CrossRef]
- Baret, P.; Béguin, C.G.; Boukhala, H.; Caris, C.; Laulhère, J.-P.; Pierre, J.-L.; Serratrice, G. O-TRENSOX: A promising water-soluble iron chelator (Both FeIII and FeII) potentally suitable for plant nutrition and iron chelation therapy. J. Am. Chem. Soc. 1995, 117, 9760–9761. [Google Scholar]
- Guo, W.; Li, J.; Fan, N.; Wu, W.; Zhou, P.; Xia, C. A simple and effective method for chemoselective esterification of phenolic acids. Synth. Commun. 2005, 35, 145–152. [Google Scholar]
- Bénard, C.; Zouhiri, F.; Normand-Bayle, M.; Danet, M.; Desmaële, D.; Leh, H.; Mouscadet, J.-F.; Mbemba, G.; Thomas, C.-M.; Bonnenfant, S.; Le Bret, M.; d’Angelo, J. Linker-Modified Quinoline Derivatives Targeting HIV-1 Integrase: Synthesis and Biological Activity. Bioorg. Med. Chem. Lett. 2004, 14, 2473–2476. [Google Scholar]
- Desmaële, D.; Zouhiri, F. Unpublished Results.
- Polanski, J.; Niedbala, H.; Musiol, R.; Podeszwa, B.; Tabak, D.; Palka, A.; Mencel, A.; Finster, J.; Mouscadet, J.-F.; Le Bret, M. Fragment based approach for the investigation of HIV-1 integrase inhibition. Lett. Drug Des. Discovery 2007, 4, 99–105. [Google Scholar] [CrossRef]
- Polanski, J.; Niedbala, H.; Musiol, R.; Podeszwa, B.; Tabak, D.; Palka, A.; Mencel, A.; Finster, J.; Mouscadet, J.-F.; Le Bret, M. 5-hydroxy-6-quinaldic acid as a novel molecular scaffold for HIV-1 integrase inhibitors. Lett. Drug Des. Discovery 2006, 3, 175–178. [Google Scholar]
- Zhuang, L.; Wai, J.S.; Embrey, M.W.; Fisher, T.E.; Egbertson, M.S.; Payne, L.S.; Guare, J.P., Jr.; Vacca, J.P; Hazuda, D.J.; Felock, P J.; Wolfe, A.L.; Stillmock, K.A.; Witmer, M.V.; Moyer, G.; Schleif, W.A.; Gabryelski, L.J.; Leonard, Y.M.; Lynch, J.J., Jr.; Michelson, S.R.; Young, S.D. Design and synthesis of 8-hydroxy-[1,6]naphthyridines as novel inhibitors of HIV-1 integrase in vitro and in infected cells. J. Med. Chem. 2003, 46, 453–456. [Google Scholar]
- Guare, J.P.; Wai, J.S.; Gomez, R.P.; Anthony, N.J.; Jolly, S.M.; Cortes, A.R.; Vacca, J.P.; Felock, P.J.; Stillmock, K.A.; Schleif, W.A.; Moyer, G.; Gabryelski, L.J.; Jin, L.X.; Chen, I.W.; Hazuda, D.J.; Young, S.D. A series of 5-aminosubstituted 4-fluorobenzyl-8-hydroxy-[1,6]naphthyridine-7-carboxamide HIV-1 integrase inhibitors. Bioorg. Med. Chem. Lett. 2006, 16, 2900–2904. [Google Scholar]
- Lee, J.Y.; Park, J.H.; Lee, S.J.; Park, H.; Lee, Y.S. Styrylquinazoline derivatives as HIV-1 integrase inhibitors. Arch. Pharm. 2002, 335, 277–282. [Google Scholar]
- Polanski, J.; Zouhiri, F.; Jeanson, L.; Desmaële, D.; d'Angelo, J.; Mouscadet, J. F.; Gieleciak, R.; Gasteiger, J.; Le Bret, M. Use of the Kohonen neural network for rapid screening of ex vivo anti-HIV activity of styrylquinolines. J. Med. Chem. 2002, 45, 4647–4654. [Google Scholar] [CrossRef]
- Zouhiri, F.; Mouscadet, J.-F.; Mekouar, K.; Desmaële, D.; Savouré, D.; Leh, H.; Subra, F.; Le Bret, M.; Auclair, C.; d'Angelo, J. Structure-activity relationships and binding mode of Styrylquinolines as potent inhibitors of HIV-1-integrase and replication of HIV-1 in cell culture. J. Med. Chem. 2000, 43, 1533–1540. [Google Scholar]
- Yoo, H.; Lee, J.Y.; Park, J.H.; Chung, B.Y.; Lee, Y.S. Synthesis of styrylbenzofuran derivatives as styrylquinoline analogues for HIV-1 integrase inhibitors. Farmaco 2003, 58, 1243–1250. [Google Scholar]
- Pearson, D.E.; Wysong, R.D.; Breder, C.V.J. The ortho bromination of phenols. J. Org. Chem. 1967, 32, 2358–2360. [Google Scholar] [CrossRef]
- Bookser, B.C. 2-Benzyloxymethyl-5-(tributylstannyl)tetrazole. A reagent for the preparation of 5-aryl- and 5-heteroaryl-1H-tetrazoles via the Stille reaction. Tetrahedron Lett. 2000, 41, 2805–2809. [Google Scholar] [CrossRef]
- Normand-Bayle, M. Synthèse et activité biologique de nouvelles styrylquinoléines inhibitrices de l’intégrase du VIH-1.
- Espeseth, A.S.; Felock, P.; Wolfe, A.; Witmer, M.; Grobler, J.; Anthony, N.; Egbertson, M.; Melamed, J.Y.; Young, S.; Hamill, T.; Cole, J.L.; Hazuda, D.J. Proc. Natl. Acad. Sci. USA 2000, 97, 11244–11249.
- Zouhiri, F.; Desmaële, D.; d'Angelo, J.; Ourevitch, M.; Mouscadet, J.-F.; Leh, H.; Le Bret, M. HIV-1 replication inhibitors of the styrylquinoline class: incorporation of a masked diketo acid pharmacophore. Tetrahedron Lett. 2001, 42, 8189–8192. [Google Scholar]
- Normand-Bayle, M.; Bénard, C.; Zouhiri, F.; Mouscadet, J.-F.; Leh, H.; Thomas, C.-M.; Mbemba, G.; Desmaële, D.; d’Angelo, J. New HIV-1 replication inhibitors of the styryquinoline class bearing aroyl/acyl groups at the C-7 position: Synthesis and biological activity. Bioorg. Med. Chem. Lett. 2005, 15, 4019–4022. [Google Scholar]
- Zouhiri, F.; Danet, M.; Bénard, C.; Normand-Bayle, M.; Mouscadet, J.-F.; Leh, H.; Thomas, C.-M.; Mbemba, G.; d'Angelo, J.; Desmaële, D. HIV-1 replication inhibitors of the styrylquinoline class: Introduction of an additional carboxyl group at the C-5 position of the quinoline. Tetrahedron Lett. 2005, 46, 2201–2205. [Google Scholar]
- Michaut, M.; Monneret, C.; Soma, E.; Thibault, L.; Wermuth, C. Styrylquinolines, their process of preparartion and their therapeutic uses. PCT Int. Appl. WO/2010/010148 A1, 2010. [Google Scholar]
- Zeng, C.; Niu, L.; Ping, D.; Zhong, R. Design and synthesis of 2-styrylquinoline-7-sulfonamide derivatives as potential HIV integrase inhibitors. Chin. J. Org. Chem. 2009, 29, 1105–1114. [Google Scholar]
- Auclair, C.; Giethlen, B.; Michaut, M.; Monneret, C.; Soma, E.; Thibault, L.; Wermuth, C. Styrylquinolines, their process of preparartion and their therapeutic uses. PCT Int. Appl. WO/2010/010147 A1, 2010. [Google Scholar]
- Chow, S.A.; Vincent, K.A.; Ellison, V.; Brown, P.O. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science 1992, 255, 723–726. [Google Scholar]
- Courcot, B.; Firley, D.; Fraisse, B.; Becker, P.; Gillet, J. M.; Pattison, P.; Chernyshov, D.; Sghaier, M.; Zouhiri, F.; Desmaele, D.; d'Angelo, J.; Bonhomme, F.; Geiger, S.; Ghermani, N.E. Crystal and electronic structures of magnesium(II), copper(II), and mixed magnesium(II)-copper(II) complexes of the quinoline half of styrylquinoline-type HIV-1 integrase inhibitors. J. Phys. Chem. 2007, B111, 6042–6050. [Google Scholar]
- Ouali, M.; Laboulais, C.; Leh, H.; Gill, D.; Desmaele, D.; Mekouar, K.; Zouhiri, F.; d'Angelo, J.; Auclair, C.; Mouscadet, J.-F.; Le Bret, M. Modeling of the inhibition of retroviral integrases by styrylquinoline derivatives. J. Med. Chem. 2000, 43, 1949–1957. [Google Scholar]
- Ouali, M.; Laboulais, C.; Leh, H.; Gill, D.; Xhuvani, E.; Zouhiri, F.; Desmaele, D.; d'Angelo, J.; Auclair, C.; Mouscadet, J.-F.; Le Bret, M. Tautomers of styrylquinoline derivatives containing a methoxy substituent: computation of their population in aqueous solution and their interaction with RSV integrase catalytic core. Acta Biochim. Pol. 2000, 47, 11–22. [Google Scholar]
- Ma, X.-h.; Zhang, X.-y.; Tan, J.-j.; Chen, W;-z; Wang, C.-x. Exploring binding mode for styrylquinoline HIV-1 integrase inhibitors using comparative molecular field analysis docking studies. Acta Pharmacol. Sin. 2004, 25, 950–958. [Google Scholar]
- Firley, D.; Courcot, B.; Gillet, J.M.; Fraisse, B.; Zouhiri, F.; Desmaele, D.; d'Angelo, J.; Ghermani, N.E. Experimental/theoretical electrostatic properties of a styrylquinoline-type HIV-1 integrase inhibitor and its progenitors. J. Phys. Chem. B 2006, 110, 537–547. [Google Scholar]
- Deprez, E.; Barbe, S.; Kolaski, M.; Leh, H.; Zouhiri, F.; Auclair, C.; Brochon, J.-C.; Le Bret, M.; Mouscadet, J.-F. Mechanism of HIV-1 integrase inhibition by styrylquinoline derivatives in vitro. Mol. Pharmacol. 2004, 65, 85–98. [Google Scholar] [CrossRef]
- Jenkins, T.M.; Esposito, D.; Engelman, A.; Craigie, R. Critical contacts between HIV-1 integrase and viral DNA identified by structure-based analysis and photo-crosslinking. EMBO J. 1997, 16, 6849–6859. [Google Scholar]
- Esposito, D.; Craigie, R. Sequence specificity of viral end DNA binding by HIV-1 integrase reveals critical regions for protein-DNA interaction. EMBO J. 1998, 17, 5832–5843. [Google Scholar]
- Harper, A.L.; Skinner, L.M.; Sudol, M.; Katzman, M. Use of patient-derived human immunodeficiency virus type 1 integrases to identify a protein residue that affects target site selection. J. Virol. 2001, 75, 7756–7762. [Google Scholar]
- Perryman, A.L.; McCammon, J.A. AutoDocking dinucleotides to the HIV-1 integrase core domain: exploring possible binding sites for viral and genomic DNA. J. Med. Chem. 2002, 45, 5624–5627. [Google Scholar]
- Hazuda, D.J.; Felock, P.; Witmer, M.; Wolfe, A.; Stillmock, K.; Grobler, J.A.; Espeseth, A.; Gabryelski, L.; Schleif, W.; Blau, C.; Miller, M.D. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 2000, 287, 646–650. [Google Scholar]
- Bonnenfant, S.; Thomas, C.M.; Vita, C.; Subra, F.; Deprez, E.; Zouhiri, F.; Desmaele, D.; d'Angelo, J.; Mouscadet, J.-F.; Leh, H. Styrylquinolines, integrase inhibitors acting prior to integration: a new mechanism of action for anti-integrase agents. J. Virol. 2004, 78, 5728–5736. [Google Scholar]
- Mousnier, A.; Leh, H.; Mouscadet, J.-F.; Dargemont, C. Nuclear import of HIV-1 integrase is inhibited in vitro by styrylquinoline derivatives. Mol. Pharmacol. 2004, 66, 783–788. [Google Scholar] [CrossRef]
- Malet, I.; Delelis, O.; Valantin, M.-A.; Montes, B.; Soulie, C.; Wirden, M.; Tchertanov, L.; Peytavin, G.; Reynes, J.; Mouscadet, J.-F.; Katlama, C.; Calvez, V.; Marcelin, A.-G. Mutations associated with failure of raltegravir treatment affect integrase sensitivity to the inhibitor in vitro. Antimicrob. Agents Chemother. 2008, 52, 1351–1358. [Google Scholar]
- Delelis, O.; Thierry, S.; Subra, F.; Simon, F.; Malet, I.; Alloui, C.; Sayon, S.; Calvez, V.; Deprez, E.; Marcelin, A.-G.; Tchertanov, L.; Mouscadet, J.-F. Impact of Y143 HIV-1 integrase mutations on resistance to raltegravir in vitro and in vivo. Antimicrob. Agents Chemother. 2010, 54, 491–501. [Google Scholar]
- Dvorin, J.D.; Bell, P.; Maul, G.G.; Yamashita, M.; Emerman, M.; Malim, M.H. Reassessment of the roles of integrase and the central DNA flap in human immunodeficiency virus type 1 nuclear import. J. Virol. 2002, 76, 12087–12096. [Google Scholar]
- Thibault, L.; Rochas, S.; Dourlat, J.; Monneret, C.; Carayon, K.; Deprez, E.; Mouscadet, J.F.; Soma, E.; Lebel-Binay, S. New integrase binding inhibitors acting in synergy with raltegravir. Antivir. Ther. 2009, 14, A29, abstract 27.. [Google Scholar]
© 2010 by the authors;
Share and Cite
Mouscadet, J.-F.; Desmaële, D. Chemistry and Structure-Activity Relationship of the Styrylquinoline-Type HIV Integrase Inhibitors. Molecules 2010, 15, 3048-3078. https://doi.org/10.3390/molecules15053048
Mouscadet J-F, Desmaële D. Chemistry and Structure-Activity Relationship of the Styrylquinoline-Type HIV Integrase Inhibitors. Molecules. 2010; 15(5):3048-3078. https://doi.org/10.3390/molecules15053048
Chicago/Turabian StyleMouscadet, Jean-François, and Didier Desmaële. 2010. "Chemistry and Structure-Activity Relationship of the Styrylquinoline-Type HIV Integrase Inhibitors" Molecules 15, no. 5: 3048-3078. https://doi.org/10.3390/molecules15053048