Experimental and Computational Studies on Non-Covalent Imprinted Microspheres as Recognition System for Nicotinamide Molecules
Abstract
:1. Introduction
2. Results and Discussion
2.1. Polymer Syntheses and Characterization
| Sample | Cross-linker EGDMA (mmol) | Solvent | Solvent Volume (mL) | T (°C) | Reaction Time (h) | Yield (%) | Microsphere size (μm) |
|---|---|---|---|---|---|---|---|
| P1 | 6.19 | CHCl3 | 40 | 60 | 20 | 59 | (2.0±0.2) |
| N-P1 | 6.19 | CHCl3 | 40 | 60 | 20 | 62 | (2.0±0.2) |
| P2 | 6.19 | MeCN | 40 | 60 | 20 | <5 | - |
| P3 | 6.19 | MeCN | 40 | 65 | 20 | 19 | (0.20±0.02) |
| P4 | 6.19 | MeCN | 30 | 65 | 20 | 22 | (0.35±0.02) |
| P5 | 6.19 | MeCN | 30 | 70 | 20 | 43 | (0.38±0.02) |
| P6 | 6.19 | MeCN | 30 | 70 | 48 | 45 | (0.34±0.02) |
| P7 | 9.3 | MeCN | 30 | 70 | 48 | 98 | (0.50±0.05) |
| N-P7 | 9.3 | MeCN | 30 | 70 | 48 | 95 | (0.50±0.05) |

2.2. Prepolymerization-complexation studies on the interactions between template and functional monomer
| Chemical shift (δ, ppm) | ||||
|---|---|---|---|---|
| Solvent | nia | nia + MAA | Δδ | |
| CDCl3 | H Amide | 5.93 | 6.51 | 0.58 |
| 6.17 | 7.00 | 0.83 | ||
| MeCN-d3 | H Amide | 6.22 | 6.60 | 0.38 |
| 6.90 | 7.03 | 0.13 | ||
2.3. Computational Studies on the Complexes Between nia and MAA
| Approach | 3 | 4 | 1:2 |
|---|---|---|---|
| RI-BLYP/def2-TZVP//RI-BLYP/SV(P) | -4.2 | -13.9 | -24.4 |
| RI-BLYP/def2-TZVP//RI-BLYP/def2-TZVP | -4.4 | -14.0 | -24.5 |
| B3LYP/def2-TZVP//RI-BLYP/SV(P) | -4.9 | -14.7 | -25.9 |
| B3LYP/def2-TZVP//RI-BLYP/SV(P) with BSSE | -4.5 | -14.1 | -24.7 |
| RI-MP2/def2-TZVPP//RI-BLYP/SV(P) | -7.0 | -15.9 | -29.0 |
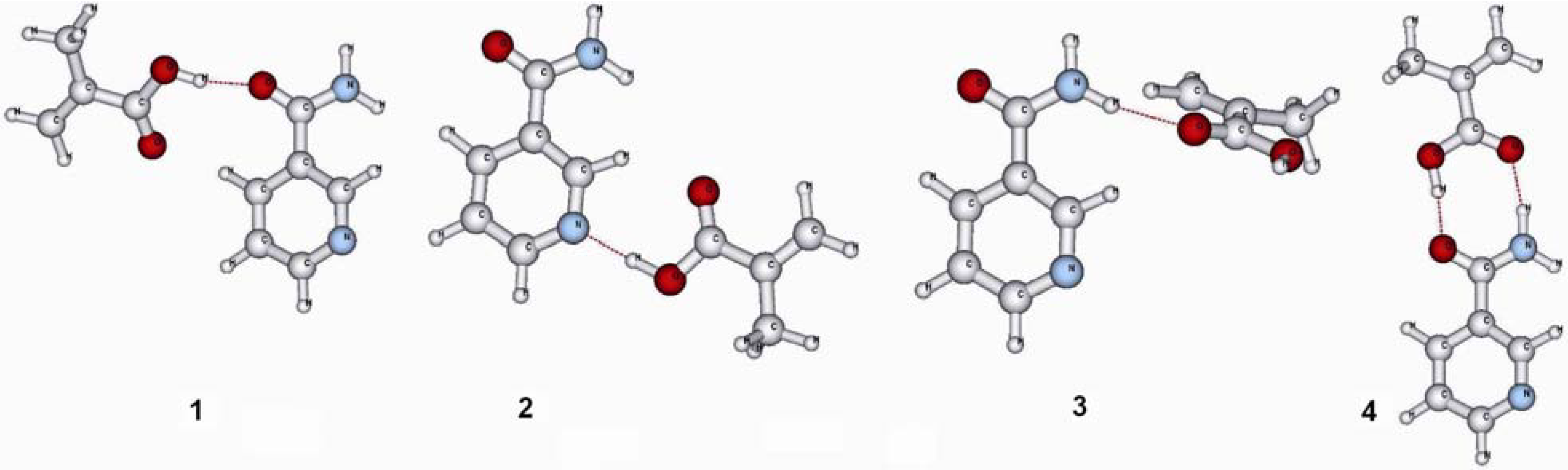
| Hydrogen bond | Gas phase | CHCl3 (ε = 4.81) | MeCN (ε = 37.5) |
|---|---|---|---|
| 1 | -9.2 (-8.7) | -5.4 (-4.9) | -4.2 (-3.8) |
| 2 | -11.5 (10.9) | -7.4 (-6.8) | -5.8 (-5.2) |
| 3 | -4.9 (-4.5) | -2.1 (-1.7) | -0.8 (-0.4) |
| 4 | -14.7 (14.1) | -9.1 (-9.5) | -6.7 (-6.1) |

| Surrounding | 1:2 | 1:3 | 1:4 |
|---|---|---|---|
| Gas phase | -25.9 | -38.5 | -46.2 |
| CHCl3 (ε = 4.81) | -16.1 | -23.0 | -27.1 |
| MeCN (ε = 37.5) | -12.3 | -16.7 | -19.9 |
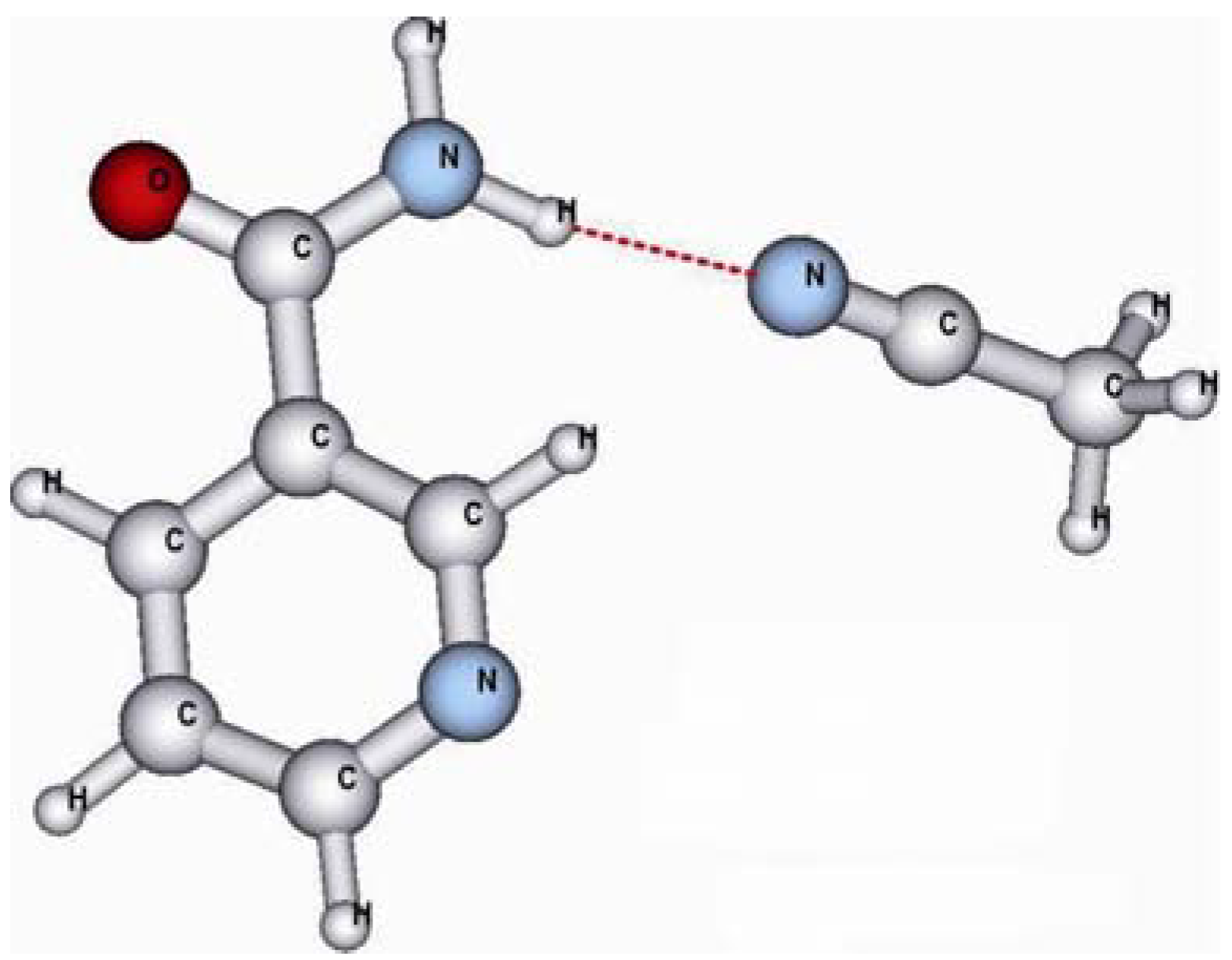
| Approach | nia-MAA | nia-MeCN |
|---|---|---|
| B3LYP/def2-TZVP (gas phase) | -4.9 | -5.4 |
| B3LYP/def2-TZVP with BSSE (gas phase) | -4.5 | -5.2 |
| B3LYP/def2-TZVP (MeCN ε = 37.5) | -0.8 | -1.2 |
| B3LYP/def2-TZVP (MeCN ε = 37.5) + BSSE | -0.4 | -1.0 |
| RI-MP2/def2-TZVPP (gas phase) | -7.0 | -7.1 |
| RI-MP2/def2-TZVPP with BSSE (gas phase) | -5.9 | -6.6 |
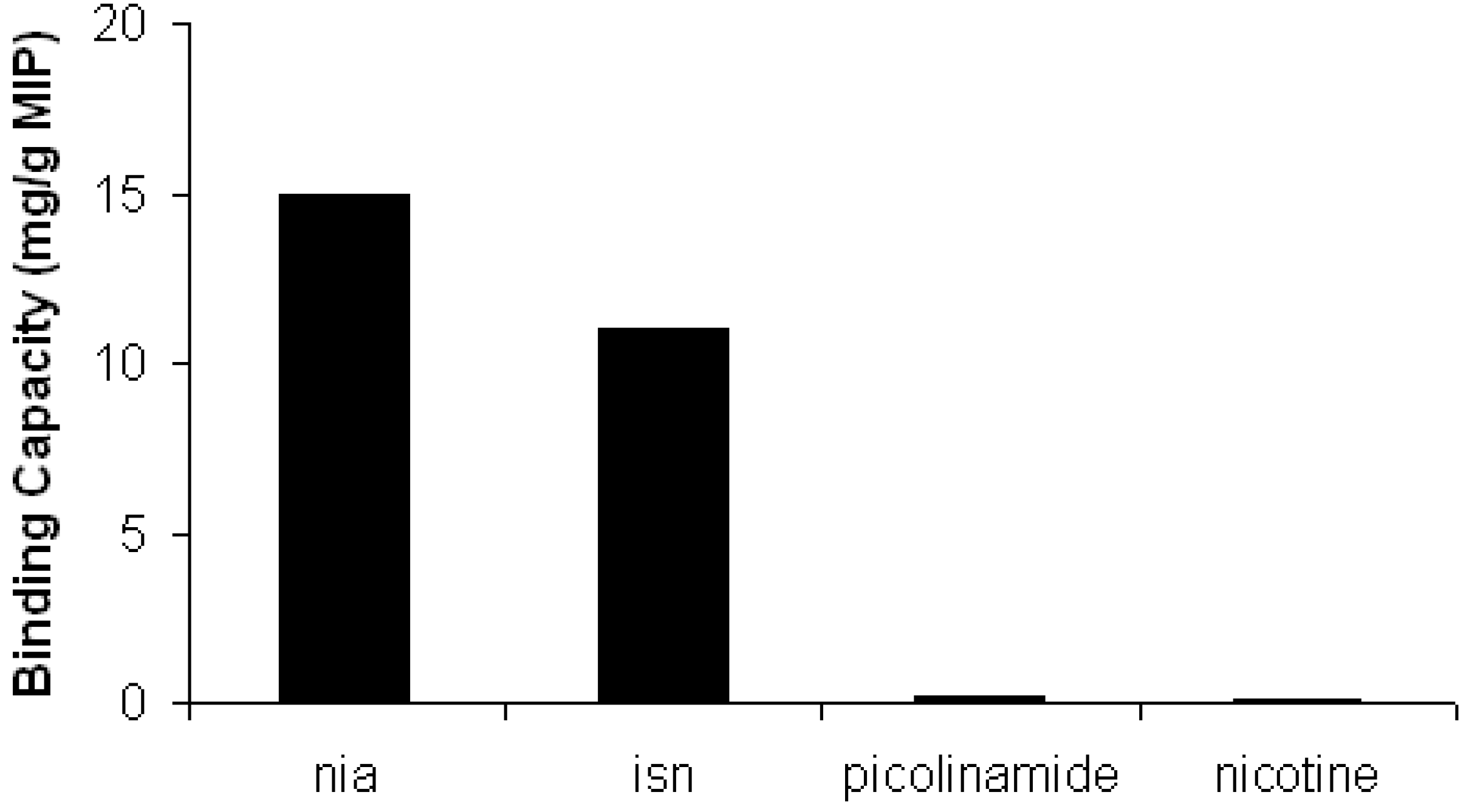
2.4. Binding Capacity Evaluation


2.5. Selectivity Evaluation
3. Conclusions
4. Experimental
4.1. General
4.2. Polymers Preparation and Template Removal
4.3. Computational Details

4.4. Calibration Curves and Binding Experiments
References
- Sellergren, B. Direct drug determination by selective sample enrichment on an imprinted polymer. Anal. Chem. 1994, 66, 1578–1582. [Google Scholar] [CrossRef]
- Sellergren, B. Polymer- and template-related factors influencing the efficiency in molecularly imprinted solid-phase extractions. Trends Anal. Chem. 1999, 18, 164–174. [Google Scholar] [CrossRef]
- Haginaka, J. Molecularly imprinted polymers for solid-phase extraction. Anal. Bioanal. Chem. 2004, 379, 332–334. [Google Scholar] [CrossRef]
- Koster, E.H.M.; Crescenzi, C.; den Hoedt, W.; Ensing, K.; de Jong, G.J. Fibers coated with molecularly imprinted polymers for solid-phase microextraction. Anal. Chem. 2001, 73, 3140–3145. [Google Scholar] [CrossRef]
- Takeuchi, T.; Haginaka, J. Separation and sensing based on molecular recognition using molecularly imprinted polymers. J. Chromatogr. B 1999, 728, 1–20. [Google Scholar] [CrossRef]
- Kempe, M.; Mosbach, K. Molecular imprinting used for chiral separations. J. Chromatogr. 1995, 694, 3–13. [Google Scholar] [CrossRef]
- Sellergren, B. Imprinted chiral stationary phases in high-performance liquid chromatography. J. Chromatogr. 2001, 906, 227–252. [Google Scholar] [CrossRef]
- Kriz, D.; Ramstrom, O.; Mosbach, K. Molecular imprinting. New possibilities for sensor technology. Anal. Chem. 1997, 69, 345A–349A. [Google Scholar] [CrossRef]
- Henry, O.Y.F.; Cullen, D.C.; Piletsky, S.A. Optical interrogation of molecularly imprinted polymers and development of MIP sensors: a review. Anal. Bioanal. Chem. 2005, 382, 947–956. [Google Scholar] [CrossRef]
- Haupt, K.; Mosbach, K. Molecularly Imprinted Polymers and Their Use in Biomimetic Sensors. Chem. Rev. 2000, 100, 2495–2504. [Google Scholar] [CrossRef]
- Tada, M.; Iwasawa, Y. Approaches to design of active structures by attaching and molecular imprinting of metal complexes on oxide surfaces. J. Mol. Catal. A: Chem. 2003, 204-205, 27–53. [Google Scholar] [CrossRef]
- Wulff, G. Templated Synthesis of Polymers - Molecularly Imprinted Materials for Recognition and Catalysis. In Templated Organic Synthesis; Diederich, F., Stang, P.J., Eds.; Wiley-VCH: Weinheim, Germany, 2000. [Google Scholar]
- Alvarez-Lorenzo, C.; Concheiro, A. Molecularly imprinted polymers for drug delivery. J. Chromatogr. B 2004, 804, 231–245. [Google Scholar] [CrossRef]
- Hilt, J.Z.; Byrne, M.E. Configurational biomimesis in drug delivery: molecular imprinting of biologically significant molecules. Adv. Drug Deliv. Rev. 2004, 56, 1599–1620. [Google Scholar] [CrossRef]
- Wulff, G.; Poll, H.G. Enzyme-analog built polymers. 23. Influence of the structure of the binding sites on the selectivity for racemic resolution. Makromol. Chem. 1987, 188, 741–748. [Google Scholar] [CrossRef]
- Wulff, G. The Covalent and Other Stoichiometric Approaches. In Molecularly Imprinted Materials; Yan, M., Ramstroem, O., Eds.; Marcel Dekker Inc: New York, NY, USA, 2005; pp. 59–92. [Google Scholar]
- Arshady, R.; Mosbach, K. Synthesis of substrate-selective polymers by host-guest polymerization. Macromol. Chem. Phys. 1981, 182, 687–692. [Google Scholar]
- Mosbach, K.; Ramstrom, O. The emerging technique of molecular imprinting and its future impact on biotechnology. Bio/Technology 1996, 14, 163–170. [Google Scholar] [CrossRef]
- Mayes, A.G.; Mosbach, K. Molecularly imprinted polymer beads: suspension polymerization using a liquid perfluorocarbon as the dispersing phase. Anal. Chem. 1996, 68, 3769–3774. [Google Scholar] [CrossRef]
- Lee, S.W.; Yang, D.H.; Kunitake, T. Regioselective imprinting of anthracenecarboxylic acids onto TiO2 gel ultrathin films: an approach to thin film sensor. Sens. Actuat. B 2005, 104, 35–42. [Google Scholar] [CrossRef]
- Li, K.; Stover, H.D.H. Synthesis of monodisperse poly(divinylbenzene) microspheres. J. Polym. Sci. Part A: Polym. Chem. 1993, 31, 3257–3263. [Google Scholar] [CrossRef]
- Turiel, E.; Martin-Esteban, A. Molecularly imprinted polymers: towards highly selective stationary phases in liquid chromatography and capillary electrophoresis. Anal. Bioanal. Chem. 2004, 378, 1876–1886. [Google Scholar] [CrossRef]
- Wang, J.; Cormack, P.A.G.; Sherrington, D.C.; Khoshdel, E. Monodisperse, molecularly imprinted polymer microspheres prepared by precipitation polymerization for affinity separation applications. Angew. Chem. Int. Ed. 2003, 42, 5336–5338. [Google Scholar] [CrossRef]
- Ye, L.; Weiss, R.; Mosbach, K. Synthesis and characterization of molecularly imprinted microspheres. Macromolecules 2000, 33, 8239–8245. [Google Scholar] [CrossRef]
- Ye, L.; Cormack, P.A.G.; Mosbach, K. Molecular imprinting on microgel spheres. Anal. Chim. Acta 2001, 435, 187–196. [Google Scholar] [CrossRef]
- Williams, A.; Ramsden, D. Nicotinamide: A double edged sword. Parkinsonism Relat. Disorders 2005, 11, 413–420. [Google Scholar] [CrossRef]
- Fu, Q.; Zheng, N.; Li, Y.Z.; Chang, W.B.; Wang, Z.M. Molecularly imprinted polymers from nicotinamide and its positional isomer. J. Mol. Recognit. 2001, 14, 151–156. [Google Scholar] [CrossRef]
- Wu, L.; Li, Y. Study on the recognition of templates and their analogues on molecularly imprinted polymer using computational and conformational analysis approaches. J. Mol. Recognit. 2004, 17, 567–574. [Google Scholar] [CrossRef]
- Wu, L.; Zhu, K.; Zhao, M.; Li, Y. Study on the recognition of templates and their analogues on molecularly imprinted polymer using computational and conformational analysis approaches. Anal. Chim. Acta. 2005, 549, 39–44. [Google Scholar] [CrossRef]
- Li, Z.; Yang, G.; Liu, S.; Chen, Y. Adsorption isotherms on nicotinamide-imprinted polymer stationary phase. J. Chromatogr. Sci. 2005, 43, 362–366. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, H.; Liao, H.; Nie, L.; Yao, S. Influence of cross-linkers amount on the performance of the piezoelectricsensor modified with molecularly imprinted polymers. Sens. Actuat. B 2005, 105, 176–182. [Google Scholar]
- Dethlefs, K.M.; Hobza, P. Ncovalent interactions: a challenge for experimental and theory. Chem. Rev. 2000, 100, 143–167. [Google Scholar] [CrossRef]
- Hohenberg, P.; Kohn, W. Inhomogeneous Electron Gas. Phys. Rev. 1964, 136, B864–B871. [Google Scholar] [CrossRef]
- Kohn, W.; Sham, L.J. Quantum density oscillations in an inhomogeneous electron gas. Phys. Rev. 1965, 137, 1697–1705. [Google Scholar] [CrossRef]
- Vahtras, O.; Almlöf, J.; Feyereisen, W. Integral approximations for LCAO-SCF calculations. Chem. Phys. Lett. 1993, 213, 514–518. [Google Scholar] [CrossRef]
- Eichkorn, K.; Treutler, O.; Öhm, H.; Häser, M.; Ahlrichs, R. Auxiliary basis sets to approximate Coulomb potentials. (Chem. Phys. Letters 240 (1995) 283). Chem. Phys. Lett. 1995, 242, 652–660. [Google Scholar] [CrossRef]
- Tsuzuki, S.; Lüthi, H.P. Interaction energies of van der Waals and hydrogen bonded systems calculated using density functional theory: assessing the PW91 model. J. Chem. Phys. 2001, 114, 3949–3957. [Google Scholar] [CrossRef]
- Jurečka, P.; Černý, J.; Hopza, P.; Salahub, D.R. Density functional theory augmented with an empirical dispersion term. Interaction energies and geometries of 80 noncovalent complexes compared with Ab Initio quantum mechanics calculations. J. Comp. Chem. 2007, 28, 555–569. [Google Scholar] [CrossRef]
- Elstner, M.; Hobza, P.; Frauenheim, T.; Suhai, S.; Kaxiras, E. Hydrogen bonding and stacking interactions of nucleic acid base pairs: a density-functional-theory based treatment. J. Chem. Phys. 2001, 114, 5149–5155. [Google Scholar] [CrossRef]
- Tuma, C.; Boese, A.D.; Handy, N.C. Predicting the binding energies of H-bonded complexes: a comparative DFT study. Phys. Chem. Chem. Phys. 1999, 1, 3939–3947. [Google Scholar] [CrossRef]
- Becke, A.D. Density-functional exchange-energy approximation with correct asymptotic behavior. Phys. Rev. A 1988, 38, 3098–3100. [Google Scholar] [CrossRef]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37, 785–789. [Google Scholar] [CrossRef]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef]
- Perdew, J.P.; Ernzerhof, M.; Burke, K. Rationale for mixing exact exchange with density functional approximations. J. Chem. Phys. 1996, 105, 9982–9985. [Google Scholar] [CrossRef]
- Perdew, J.P.; Burke, K.; Ernzerhof, M. Generalized gradient approximation made simple. Phys. Rev. Lett. 1996, 77, 3865–3868. [Google Scholar] [CrossRef]
- Ireta, J.; Neugebauer, J.; Scheffler, M. On the accuracy of DFT for describing hydrogen bonds: dependence on the bond directionality. J. Phys. Chem. A 2004, 108, 5692–5698. [Google Scholar] [CrossRef]
- Long, Q.; Ji, H.; Lv, S. DFT study on the hydrogen bonds of phenol–cyclohexanone and phenol–H2O2 in the Baeyer–Villiger oxidation. Int. J. Quant. Chem. 2009, 109, 448–458. [Google Scholar] [CrossRef]
- Boys, S.F.; Bernardi, F. The calculation of small molecular interactions by the differences of separate total energies. Some procedures with reduced errors. Mol. Phys. 1970, 19, 553–566. [Google Scholar] [CrossRef]
- Novoa, J.J.; Sosa, C. Evaluation of the density functional approximation on the computation of hydrogen bond interactions. J. Phys. Chem. 1995, 99, 15837–15845. [Google Scholar] [CrossRef]
- Hobza, P.; Šponer, J.; Reschel, T. Density functional theory and molecular clusters. J. Comp. Chem. 1995, 16, 1315–1325. [Google Scholar] [CrossRef]
- Tomura, M. Theoretical study for a complex of 1,2,5-thiadiazole with formic acid. J. Mol. Struct. (Theochem) 2008, 868, 1–5. [Google Scholar] [CrossRef]
- Del Sole, R.; De Luca, A.; Catalano, M.; Mele, G.; Vasapollo, G. Noncovalent imprinted microspheres: preparation, evaluation and selectivity of DBU template. J. Appl. Polym. Sci. 2007, 105, 2190–2197. [Google Scholar] [CrossRef]
- Nantasenamat, C.; Isarankura-Na-Ayudhya, C.; Bulow, L.; Ye, L.; Prachayasittikul, V. In silico design for synthesis of molecularly imprinted microspheres specific towards bisphenol A by precipitation polymerization. EXCLI J. 2006, 5, 103–117. [Google Scholar]
- Isarankura-Na-Ayudhya, C.; Nantasenamat, C.; Buraparuangsang, P.; Piacham, T.; Ye, L.; Bulow, L.; Prachayasittikul, V. Computational insights on sulfonamide imprinted polymers. Molecules 2008, 13, 3077–3091. [Google Scholar] [CrossRef]
- Weigend, F.; Hätting, C.; Köhn, A. Efficient use of the correlation consistent basis sets in resolution of the identity MP2 calculations. J. Chem. Phys. 2002, 116, 3175–3183. [Google Scholar] [CrossRef]
- http://avogadro.openmolecules.net/.
- Halgren, T.A. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J. Comp. Chem. 1996, 17, 490–519. [Google Scholar] [CrossRef]
- Schäfer, A.; Horn, H.; Ahlrichs, R. Fully optimized contracted Gaussian basis sets for atoms lithium to Krypton. J. Chem. Phys. 1992, 97, 2571–2577. [Google Scholar] [CrossRef]
- Ahlrichs, R.; Bär, M.; Baron, H.P.; Bauernschmitt, R.; Böcker, S.; Ehrig, M.; Eichkorn, K.; Elliott, S.; Haase, F.; Häser, M.; Horn, H.; Huber, C.; Huniar, U.; Kattannek, M.; Kölmel, C.; Kollwitz, M.; Ochsenfeld, C.; Öhm, H.; Schäfer, A.; Schneider, U.; Treutler, O.; von Arnim, M.; Weigend, F.; Weis, P.; Weiss, H. TURBOMOLE; Quant. Chem. Group, University of Karlsruhe: Karlsruhe, Germany, since 1988.
- Eichkorn, K.; Treutler, O.; Öhm, H.; Häser, M.; Ahlrichs, R. Auxiliary basis sets to approximate Coulomb potentials. Chem. Phys. Lett. 1995, 240, 283–290. [Google Scholar] [CrossRef]
- Schäfer, A.; Huber, C.; Ahlrichs, R. Fully optimized contracted Gaussian basis sets of triple zeta valence quality for atoms Li to Kr. J. Chem. Phys. 1994, 100, 5829–5835. [Google Scholar] [CrossRef]
- Klamt, A.; Schüurmann, G. COSMO: A new approach to dielectric screening in solvents with explicit expressions for the screening energy and its gradient. J. Chem. Soc. Perkin Trans. 2 1993, 5, 799–805. [Google Scholar]
- Sample Availability: Available from the authors.
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Del Sole, R.; Lazzoi, M.R.; Arnone, M.; Sala, F.D.; Cannoletta, D.; Vasapollo, G. Experimental and Computational Studies on Non-Covalent Imprinted Microspheres as Recognition System for Nicotinamide Molecules. Molecules 2009, 14, 2632-2649. https://doi.org/10.3390/molecules14072632
Del Sole R, Lazzoi MR, Arnone M, Sala FD, Cannoletta D, Vasapollo G. Experimental and Computational Studies on Non-Covalent Imprinted Microspheres as Recognition System for Nicotinamide Molecules. Molecules. 2009; 14(7):2632-2649. https://doi.org/10.3390/molecules14072632
Chicago/Turabian StyleDel Sole, Roberta, Maria Rosaria Lazzoi, Mario Arnone, Fabio Della Sala, Donato Cannoletta, and Giuseppe Vasapollo. 2009. "Experimental and Computational Studies on Non-Covalent Imprinted Microspheres as Recognition System for Nicotinamide Molecules" Molecules 14, no. 7: 2632-2649. https://doi.org/10.3390/molecules14072632




