Multivariate Generalized Multiscale Entropy Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Multiscale Entropy
- A coarse-graining procedure. For a monovariate discrete signal of length N , the coarse-grained time series is computed asFor scale one, the coarse-grained time series corresponds to the original signal. The length of the coarse-grained time series is . Coarse-graining can be seen as an averaging of the data inside a window of length τ (to reduce the high frequency components), followed by a downsampling of the averaged data by a factor τ [10]. Moreover, it has been reported that Equation (1) is similar to the use of a finite-impulse response (FIR) filter on the original time series x and to the downsampling of the filtered signal with a factor τ [11]. This FIR filter is a low-pass filter.
- Computation of the sample entropy for each coarse-grained time series. The sample entropy quantifies the regularity of finite length time series [3]. A low value for the sample entropy reflects a high degree of regularity, while a random signal has a relatively higher value of sample entropy. Sample entropy is a conditional probability measure that quantifies the likelihood that a sequence of m consecutive data points—that matches another sequence of the same length (match within a tolerance of r)—will still match the other sequence when their length is increased by one sample (sequences of length ); m therefore defines the length of the patterns that are compared to each other [3]. For a time series , the sample entropy is computed aswhere represents the total number of m-dimensional matched vector pairs, and represents the total number of -dimensional matched vector pairs. More precisely,withand is the number of vectors such that . In the definition of sample entropy, the distance d between two vectors is defined as the maximum absolute difference of their corresponding scalar components [3].
2.2. Refined Composite Multiscale Entropy
- For a scale factor τ, τ coarse-grained time series are generated. The k-th coarse-grained time series for a scale factor τ is defined as , where [12]
- For each scale factor τ, and for all τ coarse-grained time series, the number of matched vector pairs and is computed, where represents the total number of m-dimensional matched vector pairs and is computed from the k-th coarse-grained time series at a scale factor τ
- rcMSE is then defined as [5]
2.3. Multivariate (Refined Composite) Multiscale Entropy
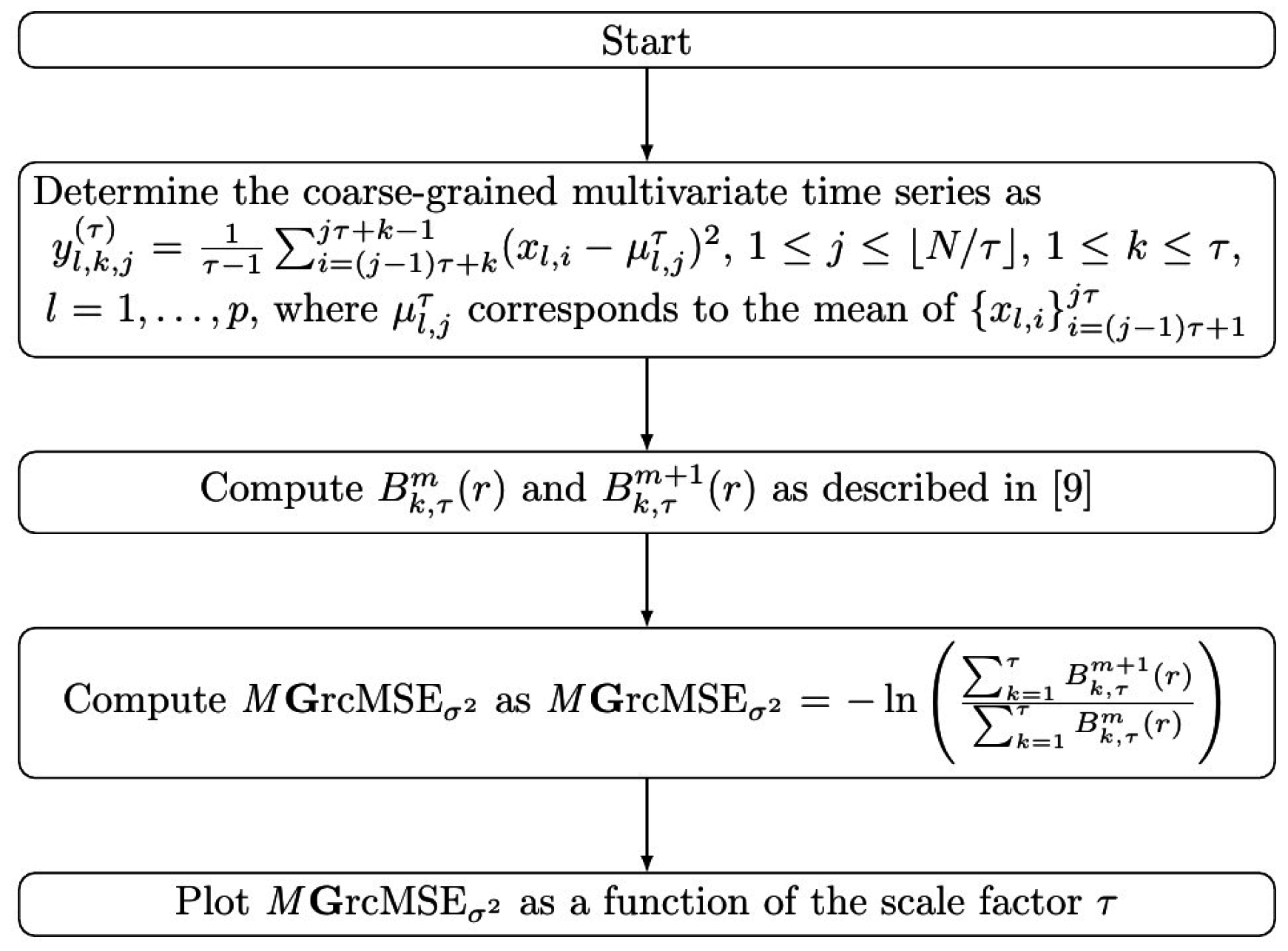
2.4. Multivariate Generalized (Refined Composite) Multiscale Entropy
2.5. Datasets Acquisition
3. Experimental Results and Discussion
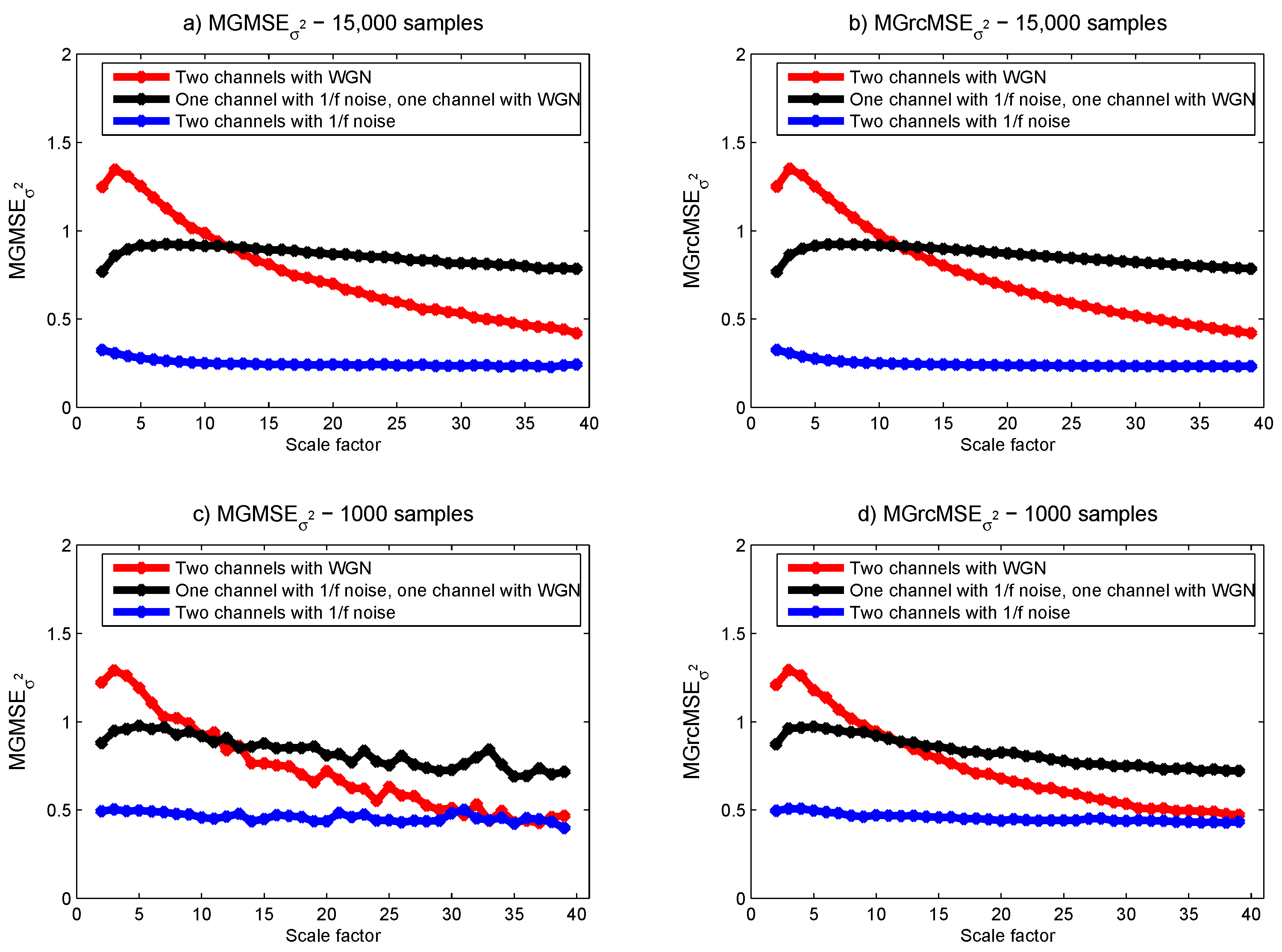
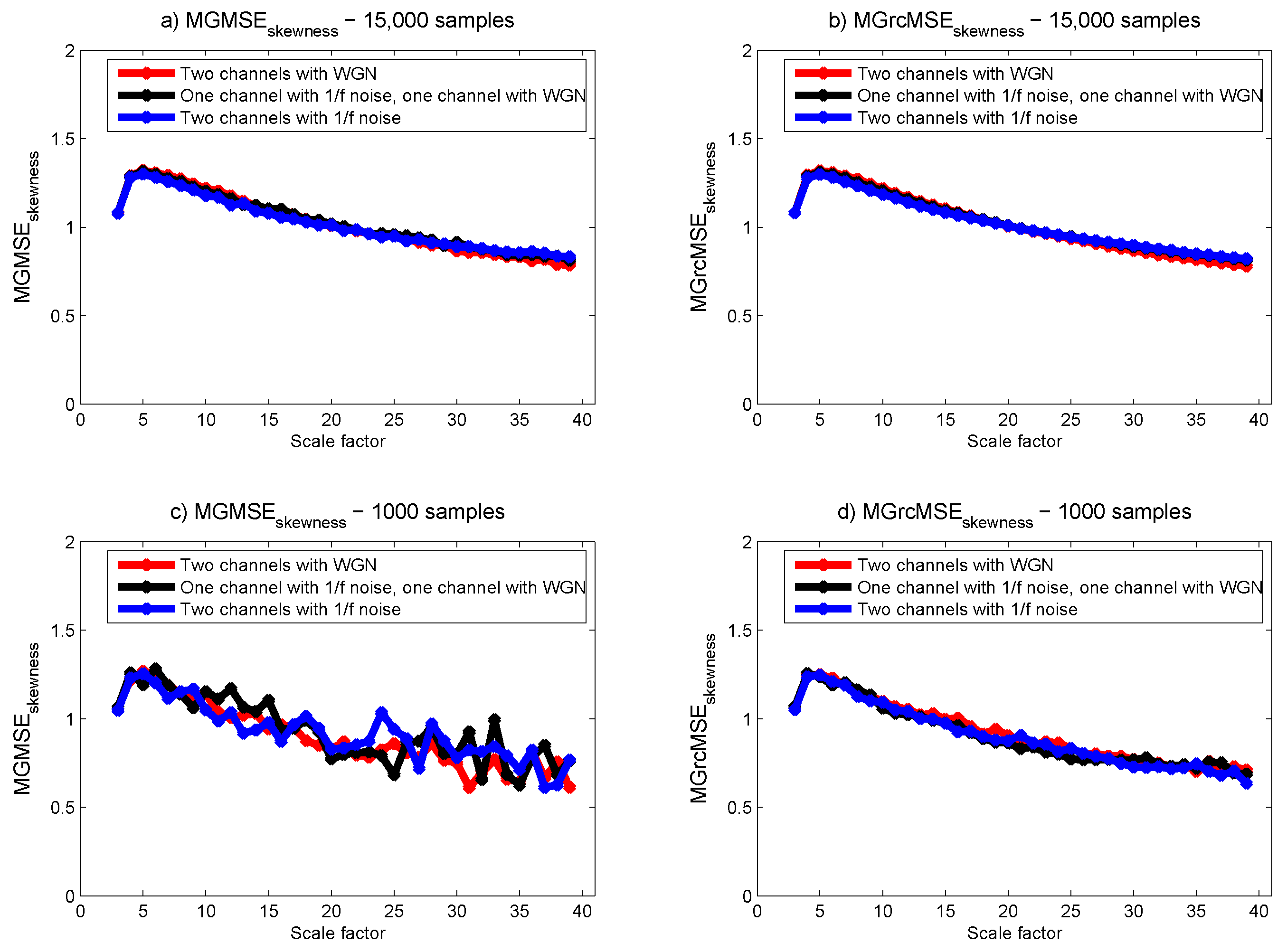
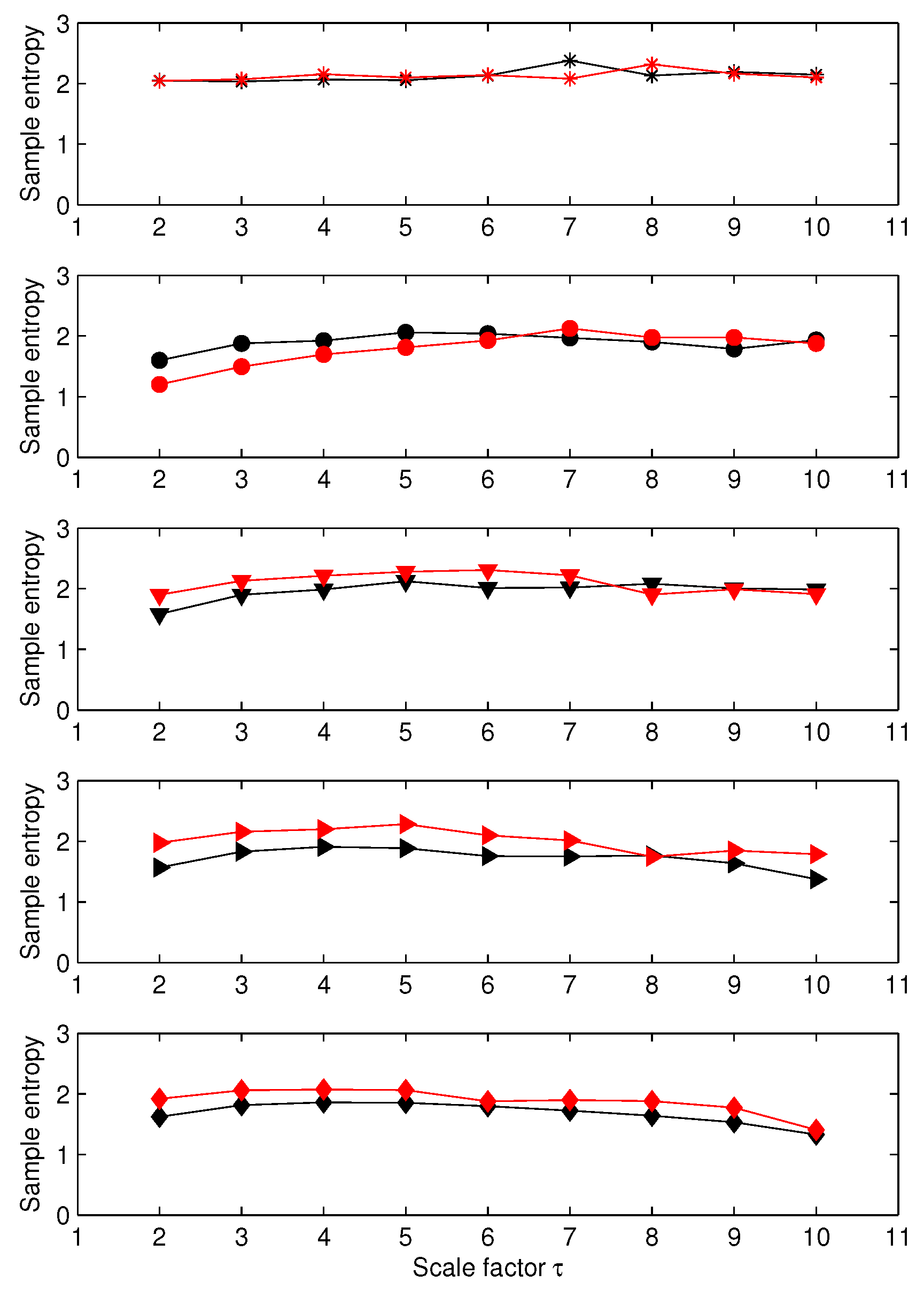
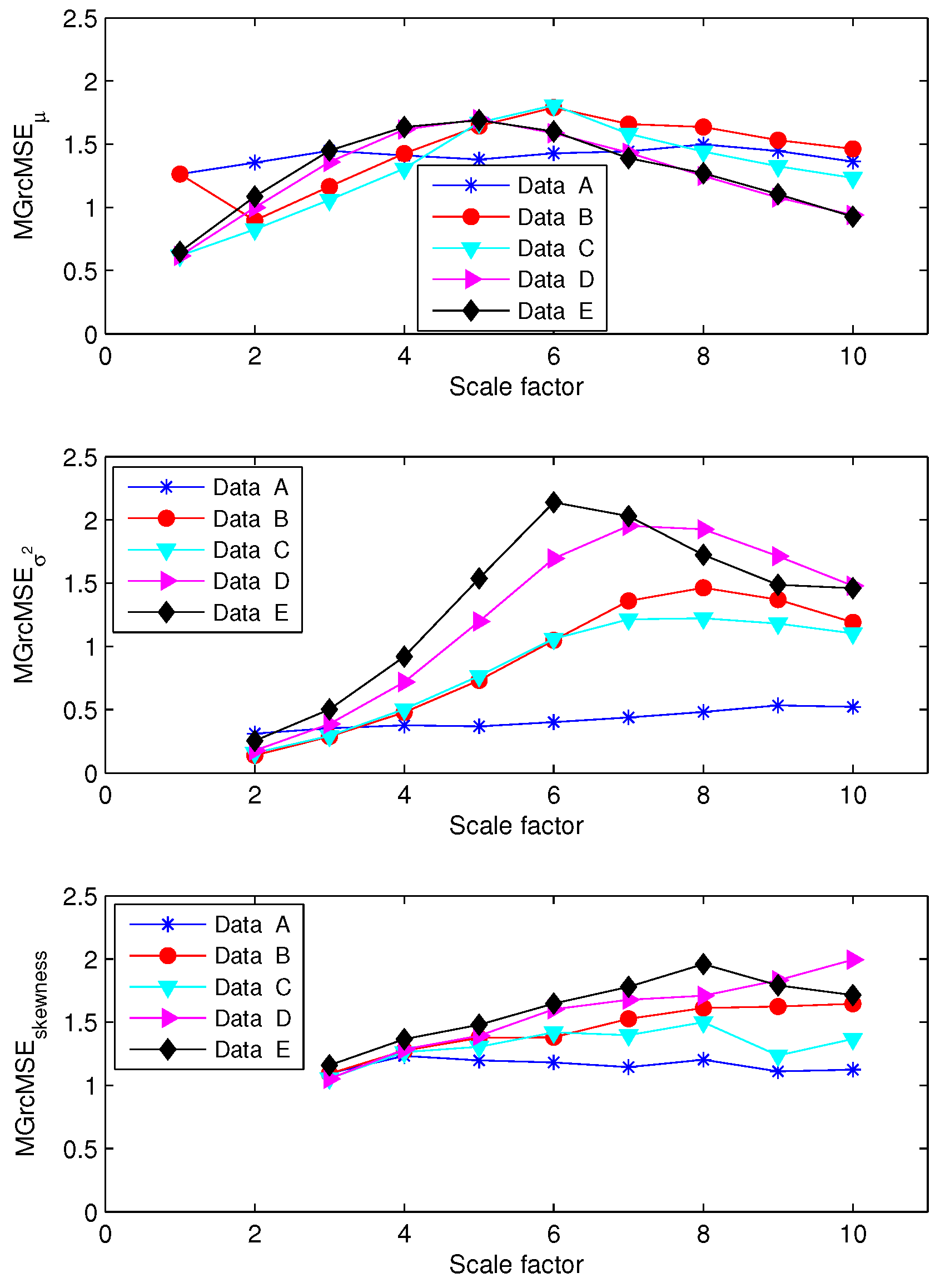
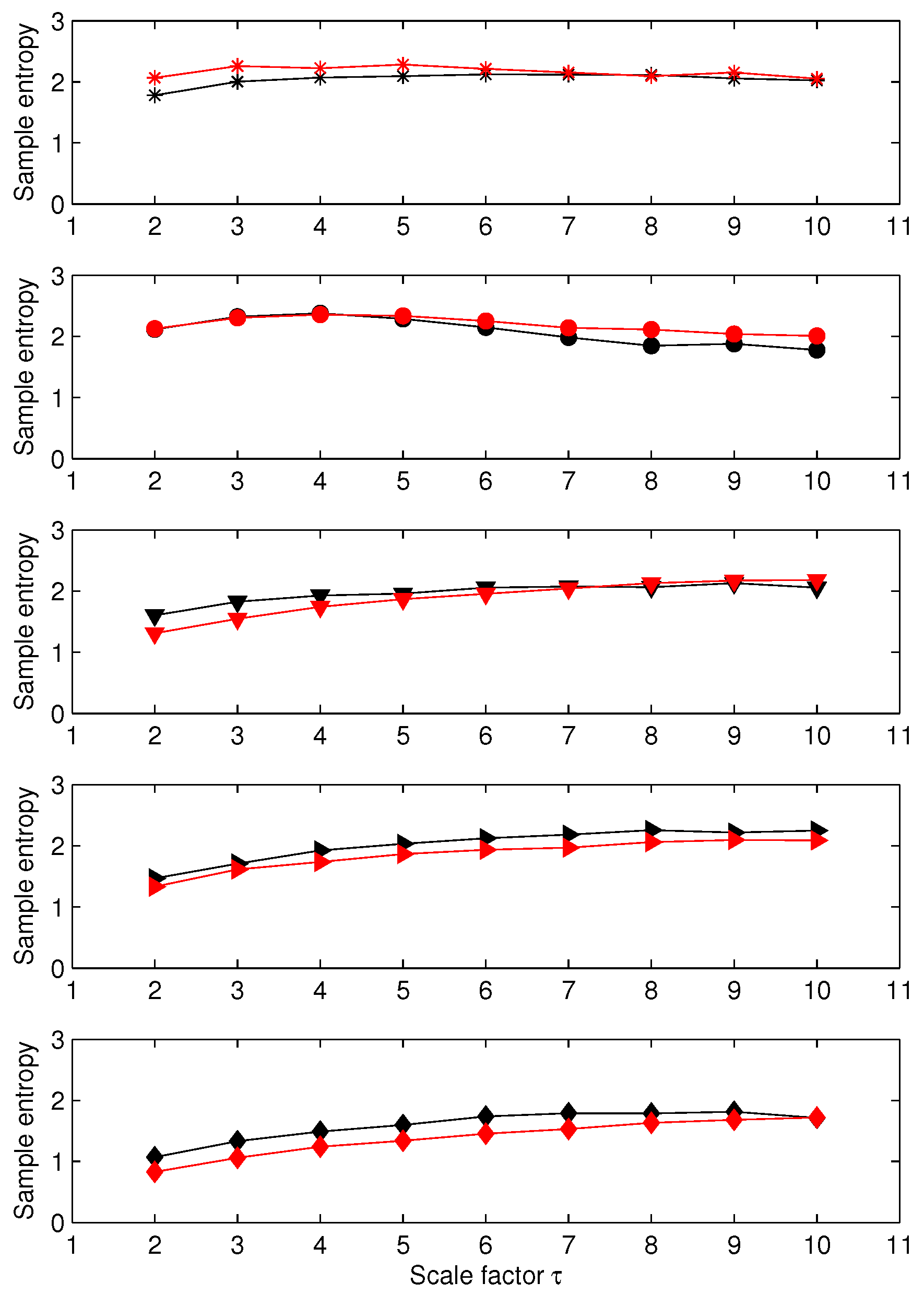
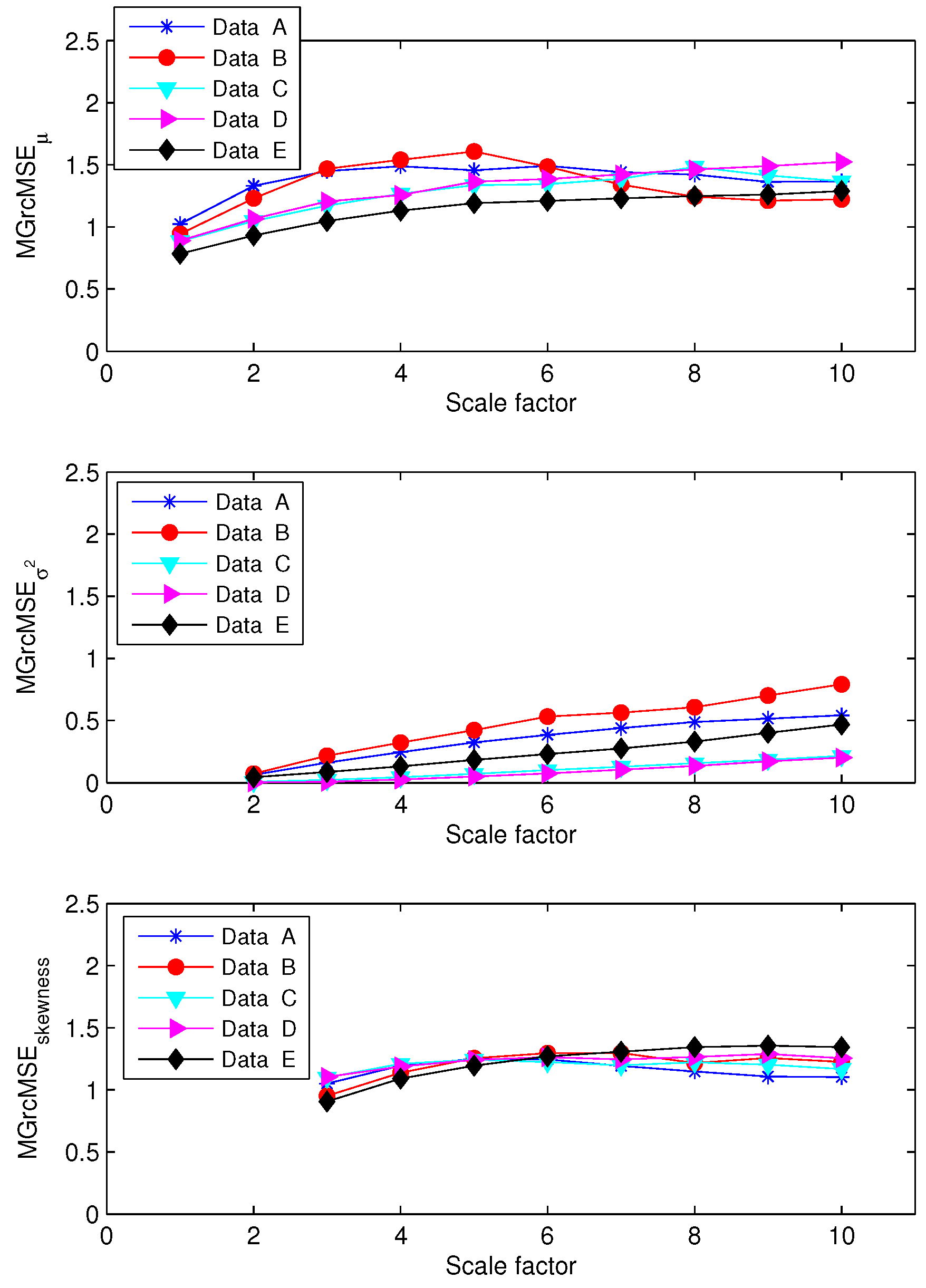
3.1. Results for Synthetic Signals
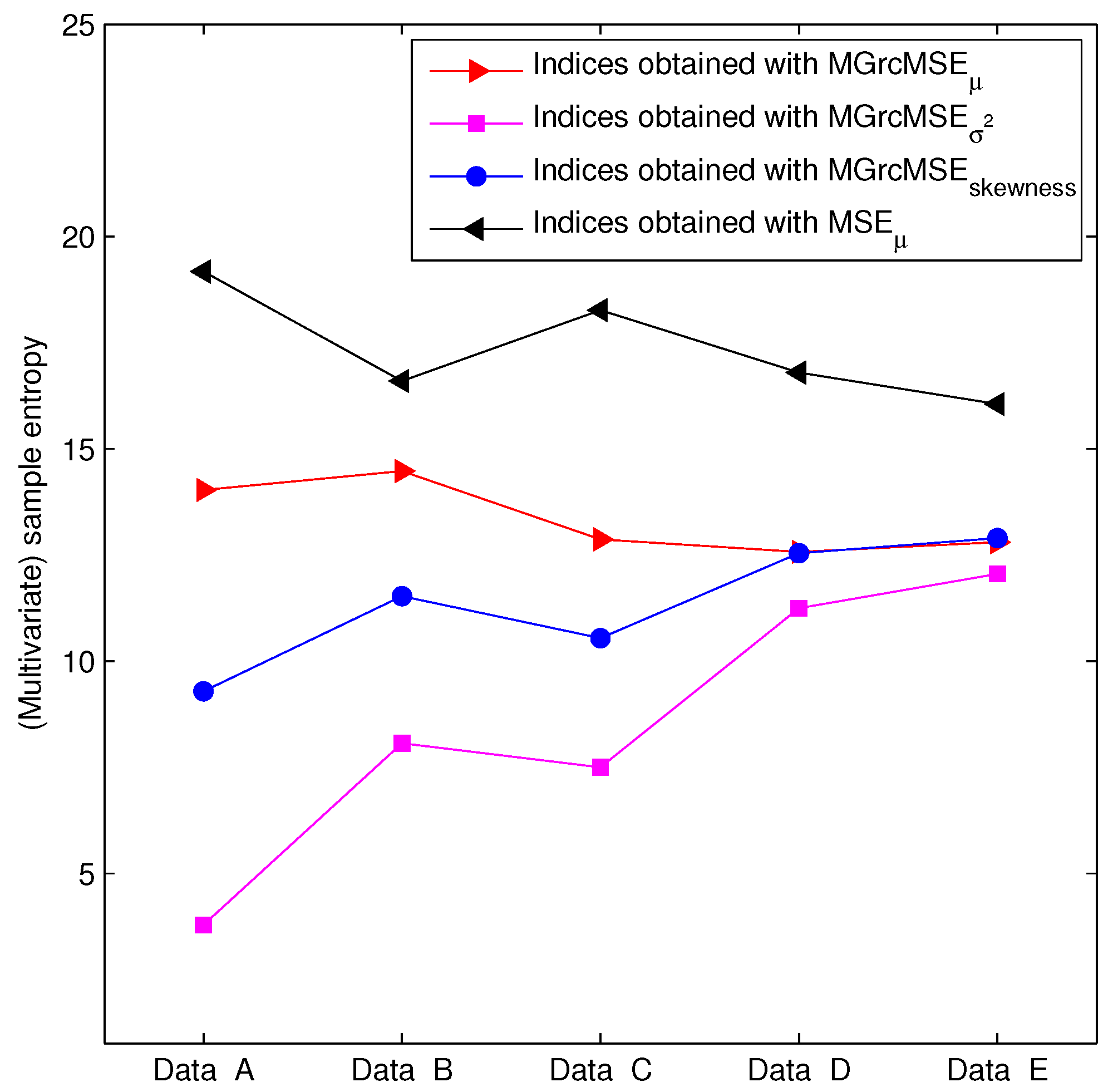
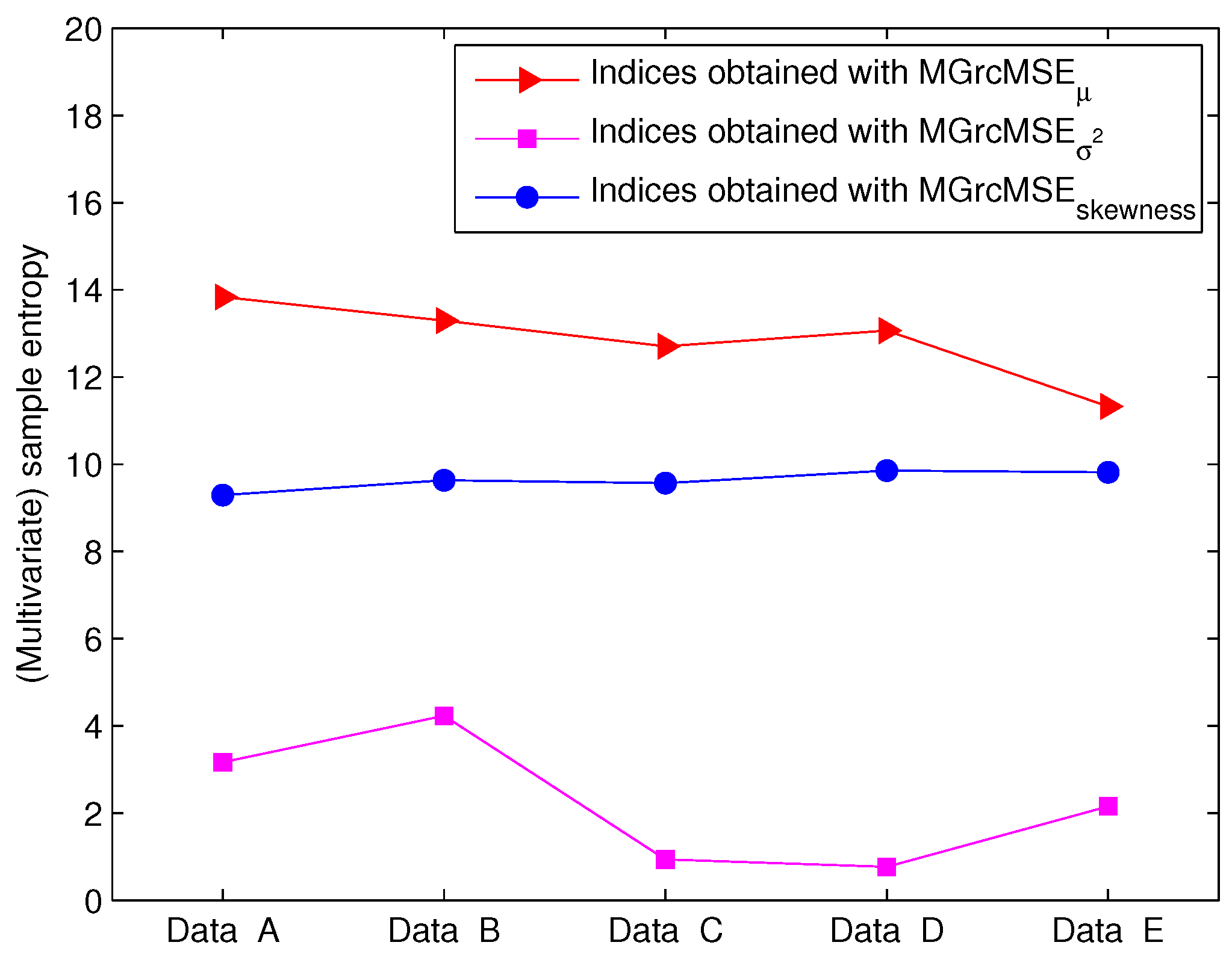
3.2. Results for Biomedical Datasets
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Costa, M.; Goldberger, A.L.; Peng, C.K. Multiscale entropy analysis of complex physiologic time series. Phys. Rev. Lett. 2002, 89, 068102. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.; Goldberger, A.L.; Peng, C.K. Multiscale entropy analysis of biological signals. Phys. Rev. E 2005, 71, 021906. [Google Scholar] [CrossRef] [PubMed]
- Richman, J.S.; Moorman, J.R. Physiological time-series analysis using approximate entropy and sample entropy. Am. J. Physiol. Heart Circ. Physiol. 2000, 278, H2039–H2049. [Google Scholar] [PubMed]
- Humeau-Heurtier, A. The multiscale entropy algorithm and its variants: A review. Entropy 2015, 17, 3110–3123. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.D.; Wu, C.W.; Lin, S.G.; Lee, K.Y.; Peng, C.K. Analysis of complex time series using refined composite multiscale entropy. Phys. Lett. A 2014, 378, 1369–1374. [Google Scholar] [CrossRef]
- Costa, M.D.; Goldberger, A.L. Generalized multiscale entropy analysis: Application to quantifying the complex volatility of human heartbeat time series. Entropy 2015, 17, 1197–1203. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.U.; Mandic, D.P. Multivariate multiscale entropy: A tool for complexity analysis of multichannel data. Phys. Rev. E 2011, 84, 061918. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.U.; Mandic, D.P. Multivariate multiscale entropy analysis. IEEE Signal Process. Lett. 2012, 19, 91–94. [Google Scholar] [CrossRef]
- Humeau-Heurtier, A. Multivariate refined composite multiscale entropy analysis. Phys. Lett. A 2016, 380, 1426–1431. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.D.; Wu, C.W.; Lee, K.Y.; Lin, S.G. Modified multiscale entropy for short-term time series analysis. Physica A 2013, 392, 5865–5873. [Google Scholar] [CrossRef]
- Valencia, J.F.; Porta, A.; Vallverdu, M.; Claria, F.; Baranowski, R.; Orlowska-Baranowska, E.; Caminal, P. Refined multiscale entropy: Application to 24-h Holter recordings of heart period variability in healthy and aortic stenosis subjects. IEEE Trans. Biomed. Eng. 2009, 56, 2202–2213. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.D.; Wu, C.W.; Lin, S.G.; Wang, C.C.; Lee, K.Y. Time series analysis using composite multiscale entropy. Entropy 2013, 15, 1069–1084. [Google Scholar] [CrossRef]
- Available online: http://www2.le.ac.uk/departments/engineering/research/bioengineering/neuroengineering-lab/software (accessed on 5 June 2016).
- Available online: http://epileptologie-bonn.de/cms/frontcontent.php?idcat=193&lang=3 (accessed on 22 October 2016).
- Quian Quiroga, R.; Kraskov, A.; Kreuz, T.; Grassberger, P. Performance of different synchronization measures in real data: A case study on electroencephalographic signals. Phys. Rev. E 2002, 65, 041903. [Google Scholar] [CrossRef] [PubMed]
- Quian Quiroga, R.; Kreuz, T.; Grassberger, P. Event synchronization: A simple and fast method to measure synchronicity and time delay patterns. Phys. Rev. E 2002, 66, 41904. [Google Scholar] [CrossRef] [PubMed]
- Andrzejak, R.G.; Lehnertz, K.; Rieke, C.; Mormann, F.; David, P.; Elger, C.E. Indications of nonlinear deterministic and finite dimensional structures in time series of brain electrical activity: Dependence on recording region and brain state. Phys. Rev. E 2001, 64, 061907. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.U.; Rehman, N.; Looney, D.; Rutkowski, T.M.; Mandic, D.P. Dynamical complexity of human responses: A multivariate data-adaptive framework. Bull. Pol. Acad. Sci. Tech. Sci. 2012, 60, 433–445. [Google Scholar] [CrossRef]
- Azami, H.; Fernandez, A.; Escudero, J. Refined Multiscale Fuzzy Entropy Based on Standard Deviation for Biomedical Signal Analysis. 2016; arXiv:1602.02847. [Google Scholar]
- Schnettler, W.T.; Goldberger, A.L.; Ralston, S.J.; Costa, M. Complexity analysis of fetal heart rate preceding intrauterine demise. Eur. J. Obstet. Gynecol. Reprod. Biol. 2016, 203, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.; Ghiran, I.; Peng, C.-K.; Nicholson-Weller, A.; Goldberger, A.L. Complex dynamics of human red blood cell flickering: Alterations with in vivo aging. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 2008, 78, 020901. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.D.; Henriques, T.; Munshi, M.N.; Segal, A.R.; Goldberger, A.L. Dynamical glucometry: Use of multiscale entropy analysis in diabetes. Chaos 2014, 24, 033139. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.D.; Schnettler, W.T.; Amorim-Costa, C.; Bernardes, J.; Costa, A.; Goldberger, A.L.; Ayres-de-Campos, D. Complexity-loss in fetal heart rate dynamics during labor as a potential biomarker of acidemia. Early Hum. Dev. 2014, 90, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Starck, J.-L.; Murtagh, F.; Fadili, J.M. Sparse Image and Signal Processing: Wavelets and Related Geometric Multiscale Analysis, 2nd ed.; Cambridge University Press: Cambridge, UK, 2015; pp. 1–423. [Google Scholar]
- Shuman, D.I.; Faraji, M.J.; Vandergheynst, P. A multiscale pyramid transform for graph signals. IEEE Trans. Signal Process. 2016, 64, 2119–2134. [Google Scholar] [CrossRef]
- Xiao, M.; He, Z. Remote sensing image fusion based on gaussian mixture model and multiresolution analysis. Proc. SPIE 2013, 8921. [Google Scholar] [CrossRef]
- Grohs, P. Geometric multiscale decompositions of dynamic low-rank matrices. Comput. Aided Geom. Des. 2013, 30, 805–826. [Google Scholar] [CrossRef]
- Sugisaki, K.; Ohmori, H. Online estimation of complexity using variable forgetting factor. In Proceedings of the 2007 SICE Annual Conference, Takamatsu, Japan, 17–20 September 2007; pp. 1–6.
- Samani, A.; Madeleine, P. Permuted sample entropy. Commun. Stat. Simul. Comput. 2010, 39, 1506–1516. [Google Scholar] [CrossRef]
- Pan, Y.H.; Lin, W.Y.; Wang, Y.H.; Lee, K.T. Computing multiscale entropy with orthogonal range search. J. Mar. Sci. Technol. 2011, 19, 107–113. [Google Scholar]
- Jiang, Y.; Mao, D.; Xu, Y. A fast algorithm for computing sample entropy. Adv. Adapt. Data Anal. 2011, 3, 167–186. [Google Scholar] [CrossRef]
- Chang, Y.C.; Wu, H.T.; Chen, H.R.; Liu, A.B.; Yeh, J.J.; Lo, M.T.; Tsao, J.H.; Tang, C.J.; Tsai, I.T.; Sun, C.K. Application of a modified entropy computational method in assessing the complexity of pulse wave velocity signals in healthy and diabetic subjects. Entropy 2014, 16, 4032–4043. [Google Scholar] [CrossRef]
- Shi, W.; Shang, P.; Ma, Y.; Sun, S.; Yeh, C.H. A comparison study on stages of sleep: Quantifying multiscale complexity using higher moments on coarse-graining. Commun. Nonlinear Sci. Numer. Simul. 2017, 44, 292–303. [Google Scholar] [CrossRef]
- Nikulin, V.V.; Brismar, T. Comment on “Multiscale entropy analysis of complex physiologic time series”. Phys. Rev. Lett. 2004, 92, 089803. [Google Scholar] [CrossRef] [PubMed]


© 2016 by the author; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Humeau-Heurtier, A. Multivariate Generalized Multiscale Entropy Analysis. Entropy 2016, 18, 411. https://doi.org/10.3390/e18110411
Humeau-Heurtier A. Multivariate Generalized Multiscale Entropy Analysis. Entropy. 2016; 18(11):411. https://doi.org/10.3390/e18110411
Chicago/Turabian StyleHumeau-Heurtier, Anne. 2016. "Multivariate Generalized Multiscale Entropy Analysis" Entropy 18, no. 11: 411. https://doi.org/10.3390/e18110411





