2.1. Overview of the Method
In a spatially extended system a source x(r) gives rise to a measurable quantity y(r′). We restrict the description to a discretized spatial lattice and consider that measurements subject to noise ξ are modeled by a linear matrix model y = Gx + ξ. For M sensors, T the length of the time series of measurements, N the number of sites of the lattice where x lives, the dimensions of the matrices y, x, G and ξ are respectively M × T, N × T, M × N and M × T. In the case of fMRI x may be taken as a variable that takes one of two values, active or inactive and G is the Bold hemodynamic response. For EEG, x represents electric dipoles and G the Green function of the brain and the y’s are scalp voltage measurements. Call the determination of y given x the forward problem. The inverse problem is to determine the sources from the data. The representation of the space where the x variables live can be done at different levels of resolution, and a set of renormalized lattices {Λd}d=0,…D is considered. Each lattice is composed by sites rd(i) ∈ Λd with i = 1, ….|Λd|. The particular form of how a coarser or renormalized lattice Λd−1 is obtained from the previous Λd depends on the particular type of problem. At each scale d the set of source variables is collectively denoted xd = {xd,i} = {xd (rd (i))} and the integration measure by dxd which includes the possibility of sums over discrete variables.
The method consists of three steps, called 1.a, 1.b and 2 respectively, to be iterated from the coarser to the finer scales, which are schematically described as follows. Choose a convenient parametric family of distributions, denoted
Q(
xd|θd), where
θd is the set of parameters for the entire lattice. For
d = 0 start with a prior
. It can be shown in simulations that this choice is not critical. Suppose now that the method has been applied and a distribution
is available. To update to the next level
, the first step 1.a uses the data to determine a posterior distribution
P (
xd−1). This requires the use of Maximum Relative Entropy methods and it might lead to a distribution not in the same parametric family as the original
. The step 1.b projects it back to the parametric space resulting in
, to be considered a posterior still in the same lattice, where parameters
incorporate information from the data. This is also a Maximum Relative Entropy step, but with the reverse Kullback-Leibler divergence, since it leads back to the parametric space. The effect of these two steps is an update on the parameters at the same resolution:
Step 2, called the Backward Renormalization Group, permits enlarging the space and leads to the required
Q(
xd|θ0d):
This completes the transition from prior at level
d − 1 to prior at level
d. In the next paragraph these steps will be described in detail.
2.2. Details of the Method
Instead of looking at the distribution of probability at a single level of resolution at each point in the iterative process the state of knowledge will be represented by a distribution
P (
xd, xd−1,
y), to be obtained from a prior distribution
Q(
xd, xd−1,
y). The method of choice is the Maximum Relative Entropy (ME) because of two reasons: (
i) when constraints are imposed and the prior already satisfies the constraints ME will result in a posterior equal to the prior and (
ii) when the results of a measurement are considered as constraints, ME gives the same results as updating with Bayes rule [
4]. The Bayesian usual update can be thought of as a special case of Maximum Entropy update.
To show (
i) we perform a simple Maximum Entropy exercise: Consider a variable
ζ that takes values
z and that
Q(
z) represents our prior state of knowledge. New information is obtained, e.g., that the expected value 〈
f(
z)〉 is known to have a particular value: 〈
f(
z)〉 =
E. The update from
Q(
z) to
P(
z) is done by maximizing
The result, after satisfying normalization is the Boltzmann-Gibbs probability density
, with
λ to be chosen to satisfy 〈
f(
z)〉
P =
E. What is the value of
λ if 〈
f(
z)〉
Q =
E, if the prior already satisfied the constraint? It is simple to see that
λ = 0 and
Z(
λ) = 1, so the reuse of old data in the from of constraints, again and again doesn’t change a density
Q(
z) that already satisfies the constraint. While this sounds trivial, it will be useful since data in Bayes updates can be written as constraints. But the reuse of data using Bayes rules should be avoided.
To prove condition (
ii) we follow [
4]. Consider the problem where a distribution
P (
x) has to be obtained, first, from the knowledge that a measurement of
Y has yielded a datum
y′; second, that prior to the inclusion of such information our knowledge of
x is codified by a distribution
Q(
x) and third, that the relation between
Y and
X is codified by a likelihood
Q(
y|x). Thus we have to determine
P (
x, y) subject to constraints ∫
dx P(
x, y) =
P (
y) =
δ (
y –
y′), by maximizing
where the Lagrange multiplier is a function λ(y) since there is a constraint for each possible value of
y.
From the resulting joint density
P(
x; y) follows the desired marginal
P(
x) = ∫
dy P(
x, y) The result is
where
is the Bayes posterior
Q(
x|y′) given by Bayes theorem, which just follows from the rules of probability. This proves that Maximum Relative Entropy as an inference engine justifies the use of Bayes theorem as the update procedure when the constraint is a datum such as knowing that a measurement of
Y turned out to give a value
y′. Maximum Relative Entropy justifies that Bayes theorem should be used in the inference process.
Going back to the spatially extended problem, consider that the problem’s solution has advanced up to a coarse level of resolution d − 1 where the degrees of freedom are xd−1. Now the data yd will be used to update the information about xd. Hence, and this is probably the most important assumption, the relevant space for inference is formed by the degrees of freedom at the two scales and the data at the level of description d: (xd, xd−1, yd).
Thus, we seek the maximization of
to obtain
P (
xd, xd−1,
yd), which in addition to normalization, is subject to
The marginal of yd is known:
, since the measured data is
.
The marginal prior Q(xd−1) belongs to a parametric family and hence shall be written as
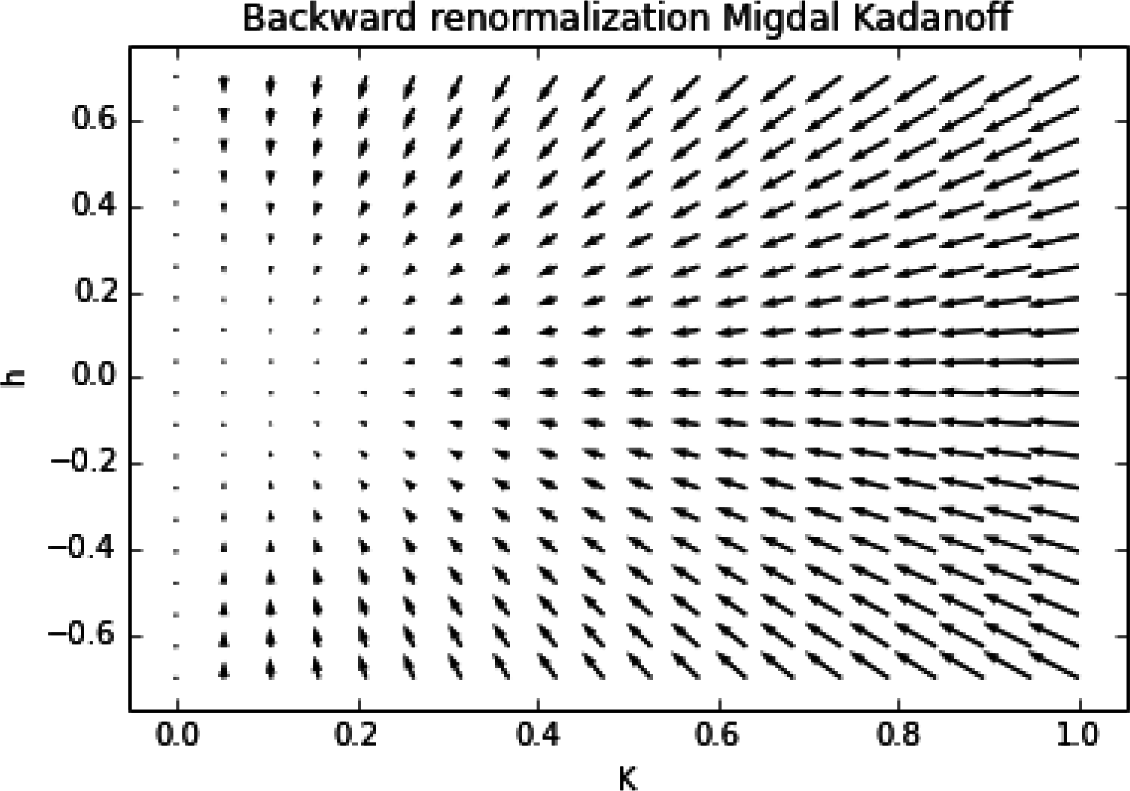
Q(xd|xd−1) codes knowledge about the backward renormalization, the relation between the x variables at different renormalization stages (see Section 3.1.)
Given xd, knowledge of xd−1 is irrelevant for yd: Q(yd|xdxd−1) = Q(yd|xd), since coarser information is unnecessary in the presence of finer information.
Marginalization and the product rule of probability give, for the prior
which can be calculated from (B) and (C). To return to the chosen parametric family, determine the closest member
by obtaining the set of
which maximizes the entropy
which is solved by imposing the partial derivatives of the entropy with respect to the parameters are zero and leads to the new parameters being given in terms of moments of
Q(
xd). This is essentially what is called a Variational Bayes step in [
5] and is a mean field approximation.
Equations (7) and
(8) implement the mapping in
Equation (2) and how to do this within the realm of the Renormalization Group is addressed in Section 3.1.
We have all the ingredients to go back to the problem of solving the maximization problem (
Equation (6)). Taking the marginal of the maximum entropy distribution we obtain
which is given by what we would have expected: Bayes theorem is used to incorporate the information in the data
through the use of the likelihood of the data
. But there is an extra and important ingredient brought in by
Equations (7) and
(8), that the prior at this new scale is obtained by whatever information we had on
xd−1,
i.e.
and by the backward renormalization procedure
Q(
xd|xd−1) that links the two scales.
Now the less dramatic point (1.b). Yet another maximum entropy projection leads to the final result of this scale resolution
which implements the mapping of equation that permits starting all over again. This step might not be necessary if the renormalization and learning steps do not lead outside the
f(
x|θ) parametric family.
Equations (9) and
(10) implement the step in display
(1).
Note that condition (ii) ensures that while we are using the measurements of yd at each scale we are not reusing the data in a forbidden manner.